4Y4E
 
 | |
4Y45
 
 | |
6O2A
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein NDN and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
1NN4
 
 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
2VOM
 
 | | Structural basis of human triosephosphate isomerase deficiency. Mutation E104D and correlation to solvent perturbation. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, de Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
6NR8
 
 | | hTRiC-hPFD Class6 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6O0Y
 
 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2VO9
 
 | | CRYSTAL STRUCTURE OF THE ENZYMATICALLY ACTIVE DOMAIN OF THE LISTERIA MONOCYTOGENES BACTERIOPHAGE 500 ENDOLYSIN PLY500 | | Descriptor: | L-ALANYL-D-GLUTAMATE PEPTIDASE, SULFATE ION, ZINC ION | | Authors: | Korndoerfer, I.P, Kanitz, A, Skerra, A. | | Deposit date: | 2008-02-09 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of the L-Alanoyl-D-Glutamate Endopeptidase Domain of Listeria Bacteriophage Endolysin Ply500 Reveals a New Member of the Las Peptidase Family.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6O25
 
 | | Crystal structure of 3945 Fab in complex with circumsporozoite protein NANP3 and anti-Kappa VHH domain | | Descriptor: | 3945 Fab heavy chain, 3945 Kappa light chain, Anti-Kappa VHH domain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
8AFR
 
 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
2W53
 
 | | Structure of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. | | Descriptor: | REPRESSOR, SULFATE ION | | Authors: | Mate, M.J, Romero, A, Hernandez, A, Martinez, J.L. | | Deposit date: | 2008-12-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Smet, the Repressor of the Stenotrophomonas Maltophilia Multidrug Efflux Pump Smedef.
J.Biol.Chem., 284, 2009
|
|
6OVN
 
 | | Crystal structure of the unliganded Clone 2 TCR | | Descriptor: | Alpha chain Clone 2 TCR, Beta chain Clone 2 TCR, CHLORIDE ION, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A TCR beta-Chain Motif Biases toward Recognition of Human CD1 Proteins.
J Immunol., 203, 2019
|
|
2WF1
 
 | | Human BACE-1 in complex with 7-ethyl-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl(methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-3,4- dihydro-1H-(1,2,5)thiadiazepino(3,4,5-hi)indole-9-carboxamide 2,2- dioxide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-7-ethyl-1-methyl-3,4-dihydro-1H-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide 2,2-dioxide | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2WF4
 
 | | Human BACE-1 in complex with 6-ethyl-1-methyl-N-((1S)-2-oxo-1-(phenylmethyl)-3-(tetrahydro-2H-pyran-4-ylamino)propyl)-1,3,4,6- tetrahydro(1,2)thiazepino(5,4,3-cd)indole-8-carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-[(1S)-1-BENZYL-2,2-DIHYDROXY-3-(TETRAHYDRO-2H-PYRAN-4-YLAMINO)PROPYL]-6-ETHYL-1-METHYL-1,3,4,6-TETRAHYDRO[1,2]THIAZEPINO[5,4,3-CD]INDOLE-8-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Howes, C, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-12 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 3: Towards Non Hydroxyethylamine Transition State Mimetics.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6OY1
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-26 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6O4W
 
 | | Binary complex of native hAChE with Donepezil | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Gerlits, O, Kovalevsky, A, Radic, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies.
Chem.Biol.Interact., 309, 2019
|
|
6O52
 
 | |
6OY4
 
 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
6O6N
 
 | | Structure of the regulator FasR from Mycobacterium tuberculosis in complex with C20-CoA | | Descriptor: | Arachinoyl-CoA, CHLORIDE ION, TetR family transcriptional regulator | | Authors: | Larrieux, N, Trajtenberg, F, Lara, J, Gramajo, H, Buschiazzo, A. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mycobacterium tuberculosis FasR senses long fatty acyl-CoA through a tunnel and a hydrophobic transmission spine.
Nat Commun, 11, 2020
|
|
6OYF
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
2WF2
 
 | | Human BACE-1 in complex with 8-ethyl-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-1-methyl-3,4,7, 8-tetrahydro-1H,6H-(1,2,5)thiadiazepino(5,4,3-de)quinoxaline-10- carboxamide 2,2-dioxide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-8-ETHYL-1-METHYL-3,4,7,8-TETRAHYDRO-1H,6H-[1,2,5]THIADIAZEPINO[5,4,3-DE]QUINOXALINE-10-CARBOXAMIDE 2,2-DIOXIDE | | Authors: | Charrier, N, Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Second Generation of Bace-1 Inhibitors Part 2: Optimisation of the Non-Prime Side Substituent.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6ETN
 
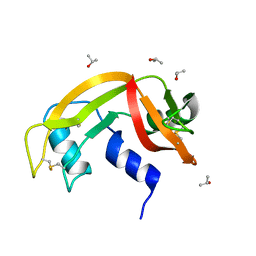 | |
1NNX
 
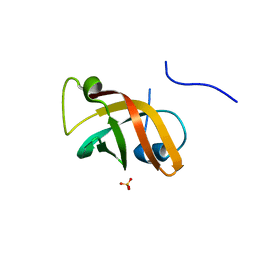 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
5JQ0
 
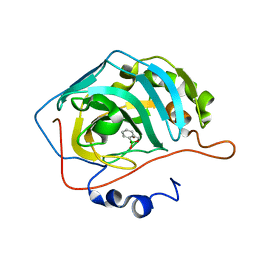 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH=8.7 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
2IDN
 
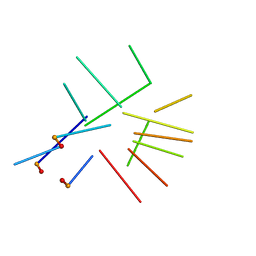 | | NMR structure of a new modified Thrombin Binding Aptamer containing a 5'-5' inversion of polarity site | | Descriptor: | 3'-D(P*GP*G*T)-5'-5'-D(P*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3' | | Authors: | Randazzo, A, Martino, L, Virno, A, Mayol, L, Giancola, C. | | Deposit date: | 2006-09-15 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site.
Nucleic Acids Res., 34, 2006
|
|
