6Z8Y
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z9N
 
 | | Copper transporter OprC | | Descriptor: | COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z9Y
 
 | | Copper transporter OprC | | Descriptor: | COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z91
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z99
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6TYM
 
 | | KEAP1 Kelch domain in complex with Compound 9 | | Descriptor: | (3S)-3-[2-(benzenecarbonyl)-5-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl]-3-(1-ethyl-4-methyl-1H-benzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.422 Å) | | Cite: | Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6TYP
 
 | | KEAP1 Kelch domain in complex with Compound 2 | | Descriptor: | (3S)-3-[2-(benzenecarbonyl)-1,2,3,4-tetrahydroisoquinolin-7-yl]-3-(1-ethyl-4-methyl-1H-benzotriazol-5-yl)propanoic acid, FORMIC ACID, Kelch-like ECH-associated protein 1 | | Authors: | Marcotte, D.J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and identification of novel, orally bioavailable non-covalent Nrf2 activators.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4R0I
 
 | | CRYSTAL STRUCTURE of MATRIPTASE in COMPLEX WITH INHIBITOR | | Descriptor: | 3-({(2S)-3-[4-(2-aminoethyl)piperidin-1-yl]-2-[(naphthalen-2-ylsulfonyl)amino]-3-oxopropyl}oxy)benzenecarboximidamide, SERINE PROTEASE, MATRIPTASE, ... | | Authors: | Rao, K.N, Ashok, K.N, Chakshusmathi, G, Rajeev, G, Subramanya, H. | | Deposit date: | 2014-07-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8HEV
 
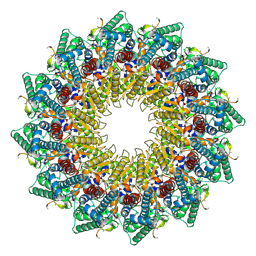 | | C12 portal in HCMV B-capsid | | Descriptor: | Portal protein, Unknown peptide | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEU
 
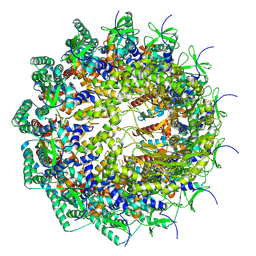 | | C12 portal in HCMV A-capsid | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
7LA4
 
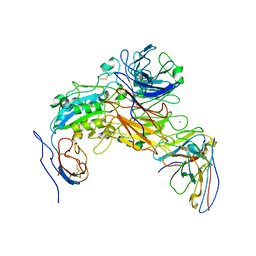 | | Integrin AlphaIIbBeta3-PT25-2 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bush, M.W, Walz, T, Coller, B, Filizola, M, Spasic, A, Nesic, D, Li, J. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron microscopy shows that binding of monoclonal antibody PT25-2 primes integrin alpha IIb beta 3 for ligand binding.
Blood Adv, 5, 2021
|
|
3T4H
 
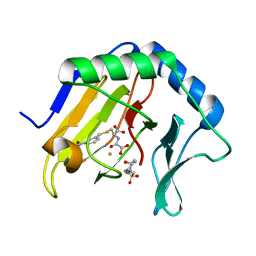 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(3-nitrobenzyl)-L-cysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, ... | | Authors: | Ma, J, Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
6D2J
 
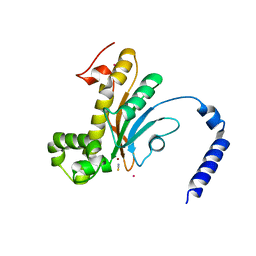 | | Beta Carbonic anhydrase in complex with thiocyanate | | Descriptor: | Carbonic anhydrase, POTASSIUM ION, THIOCYANATE ION, ... | | Authors: | Murray, A, Aggarwal, M, Pinard, M, McKenna, R. | | Deposit date: | 2018-04-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Mapping of Anion Inhibitors to beta-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa.
ChemMedChem, 13, 2018
|
|
5HJS
 
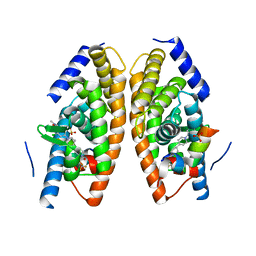 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
6UXY
 
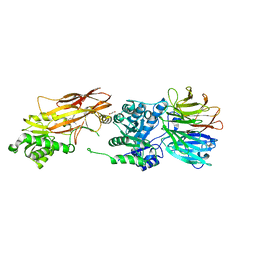 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 8 | | Descriptor: | (5R)-2-amino-5-(2-cyclohexylethyl)-3-methyl-5-phenyl-3,5-dihydro-4H-imidazol-4-one, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
2X03
 
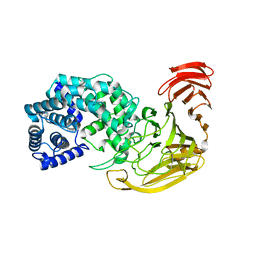 | | The X-ray structure of the Streptomyces coelicolor A3 Chondroitin AC Lyase Y253A mutant | | Descriptor: | MAGNESIUM ION, PUTATIVE SECRETED LYASE | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-12-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
8HLG
 
 | | Crystal structure of MoaE | | Descriptor: | Molybdenum cofactor biosynthesis protein D/E, SULFATE ION | | Authors: | Cai, J, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MoaE Is Involved in Response to Oxidative Stress in Deinococcus radiodurans.
Int J Mol Sci, 24, 2023
|
|
5HJP
 
 | | Identification of LXRbeta selective agonists for the treatment of Alzheimer's Disease | | Descriptor: | 2-chloro-4-{1'-[(2R)-2-hydroxy-3-methyl-2-(trifluoromethyl)butanoyl]-4,4'-bipiperidin-1-yl}-N,N-dimethylbenzamide, DI(HYDROXYETHYL)ETHER, Oxysterols receptor LXR-beta, ... | | Authors: | Parthasarathy, G, Klein, D. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification and in Vivo Evaluation of Liver X Receptor beta-Selective Agonists for the Potential Treatment of Alzheimer's Disease.
J.Med.Chem., 59, 2016
|
|
6J6Y
 
 | | FGFR4 D2 - Fab complex | | Descriptor: | Fab Heavy chain, Fab light chain, Fibroblast growth factor receptor 4 | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Preclinical Development of U3-1784, a Novel FGFR4 Antibody Against Cancer, and Avoidance of Its On-target Toxicity.
Mol.Cancer Ther., 18, 2019
|
|
8FD8
 
 | | human 15-PGDH with NADH bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-12-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
8HEY
 
 | | One CVSC-binding penton vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
8HEX
 
 | | C5 portal vertex in HCMV B-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Major capsid protein, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus.
Nat Commun, 14, 2023
|
|
2F61
 
 | | Crystal structure of partially deglycosylated acid beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid beta-glucosidase, ... | | Authors: | Hegde, R.S, Grabowski, G. | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analyses of Variant Acid beta-Glucosidases: EFFECTS OF GAUCHER DISEASE MUTATIONS.
J.Biol.Chem., 281, 2006
|
|
4IXH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-25 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Optimization of Benzoxazole-Based Inhibitors of Cryptosporidium parvum Inosine 5'-Monophosphate Dehydrogenase.
J.Med.Chem., 56, 2013
|
|
3T4V
 
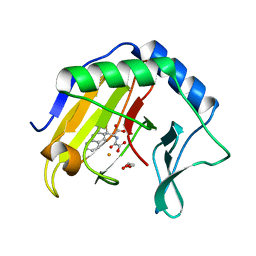 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(2-napthalenemethyl)-L-cysteine | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, GLYCEROL, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
