1TUC
 
 | |
1SWA
 
 | | APO-CORE-STREPTAVIDIN AT PH 4.5 | | Descriptor: | STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Klumb, L, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-03-02 | | Release date: | 1998-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of the streptavidin binding loop.
Protein Sci., 6, 1997
|
|
1TO9
 
 | |
1TOO
 
 | | Interleukin 1B Mutant F146W | | Descriptor: | Interleukin-1 beta | | Authors: | Adamek, D.H, Guerrero, L, Caspar, D.L. | | Deposit date: | 2004-06-14 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and energetic consequences of mutations in a solvated hydrophobic cavity.
J.Mol.Biol., 346, 2005
|
|
1TP0
 
 | |
1TVL
 
 | |
1TT7
 
 | |
1TQE
 
 | | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | | Descriptor: | Histone deacetylase 9, MEF2 binding site of nur77 promoter, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Han, A, He, J, Wu, Y, Liu, J.O. | | Deposit date: | 2004-06-17 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Recruitment of Class II Histone Deacetylases by Myocyte Enhancer Factor-2.
J.Mol.Biol., 345, 2005
|
|
1TO3
 
 | |
1TOB
 
 | | SACCHARIDE-RNA RECOGNITION IN AN AMINOGLYCOSIDE ANTIBIOTIC-RNA APTAMER COMPLEX, NMR, 7 STRUCTURES | | Descriptor: | 1,3-DIAMINO-5,6-DIHYDROXYCYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Suri, A.K, Fiala, R, Patel, D.J. | | Deposit date: | 1996-12-12 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex.
Chem.Biol., 4, 1997
|
|
1TP7
 
 | | Crystal Structure of the RNA-dependent RNA Polymerase from Human Rhinovirus 16 | | Descriptor: | 3-[BENZYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, Genome polyprotein, SULFATE ION | | Authors: | Appleby, T.C, Luecke, H, Shim, J.H, Wu, J.Z, Cheney, I.W, Zhong, W, Vogeley, L, Hong, Z, Yao, N. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer.
J.Virol., 79, 2005
|
|
1TQF
 
 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
1UHL
 
 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
1URC
 
 | | Cyclin A binding groove inhibitor Ace-Arg-Lys-Leu-Phe-Gly | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, PEPTIDE INHIBITOR | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-10-28 | | Release date: | 2003-10-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, biological activity and structural analysis of cyclic peptide inhibitors targeting the substrate recruitment site of cyclin-dependent kinase complexes.
Org. Biomol. Chem., 2, 2004
|
|
1UZV
 
 | | High affinity fucose binding of Pseudomonas aeruginosa lectin II: 1.0 A crystal structure of the complex | | Descriptor: | CALCIUM ION, PSEUDOMONAS AERUGINOSA LECTIN II, SULFATE ION, ... | | Authors: | Mitchell, E, Sabin, C.D, Snajdrova, L, Budova, M, Perret, S, Gautier, C, Gilboa-Garber, N, Koca, J, Wimmerova, M, Imberty, A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High Affinity Fucose Binding of Pseudomonas Aeruginosa Lectin Pa-Iil: 1.0 A Resolution Crystal Structure of the Complex Combined with Thermodynamics and Computational Chemistry Approaches.
Proteins, 58, 2005
|
|
1UMO
 
 | | The crystal structure of cytoglobin: the fourth globin type discovered in man | | Descriptor: | CYTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-08-26 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination.
J.Mol.Biol., 336, 2004
|
|
1TZ7
 
 | | Aquifex aeolicus amylomaltase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-alpha-glucanotransferase | | Authors: | Barends, T.R.M, Korf, H, Kaper, T, van der Maarel, M.J.E.C, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2004-07-09 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural influences on product specificity in amylomaltase from Aquifex aeolicus
TO BE PUBLISHED
|
|
1TTZ
 
 | | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris | | Descriptor: | conserved hypothetical protein | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Ho, C.K, Cooper, B, Ma, L.-C, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | X-ray structure of Northeast Structural Genomics target protein XcR50 from X. campestris
To be Published
|
|
1TWL
 
 | | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001 | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Zhou, W, Tempel, W, Liu, Z.-J, Chen, L, Clancy Kelley, L.-L, Dillard, B.D, Hopkins, R.C, Arendall III, W.B, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-07-01 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Inorganic pyrophosphatase from Pyrococcus furiosus Pfu-264096-001
To be published
|
|
1U5L
 
 | | Solution Structure of the turtle prion protein fragment (121-226) | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Calzolai, L, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of chicken, turtle, and frog
PROC.NATL.ACAD.SCI.USA, 102, 2005
|
|
1TWR
 
 | |
1U5J
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit, Met186Ile | | Descriptor: | COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2004-07-27 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1U9M
 
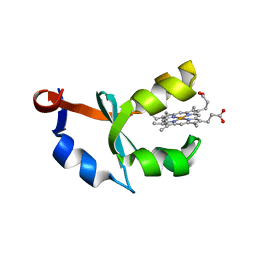 | | Crystal structure of F58W mutant of cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shan, L, Lu, J.-X, Gan, J.-H, Wang, Y.-H, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2004-08-10 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the F58W mutant of cytochrome b5: the mutation leads to multiple conformations and weakens stacking interactions.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UG3
 
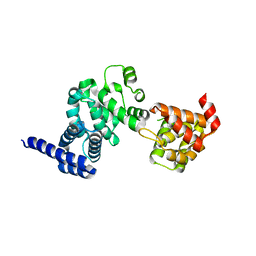 | | C-terminal portion of human eIF4GI | | Descriptor: | eukaryotic protein synthesis initiation factor 4G | | Authors: | Bellsolell, L, Cho-Park, P.F, Poulin, F, Sonenberg, N, Burley, S.K. | | Deposit date: | 2003-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1
Structure, 14, 2006
|
|
1TMX
 
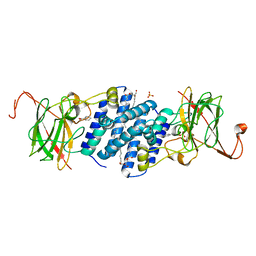 | | Crystal structure of hydroxyquinol 1,2-dioxygenase from Nocardioides Simplex 3E | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Travkin, V.M, Seifert, J, Schlomann, M, Golovleva, L, Scozzafava, A, Briganti, F. | | Deposit date: | 2004-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation.
J.Biol.Chem., 280, 2005
|
|
