8YFQ
 
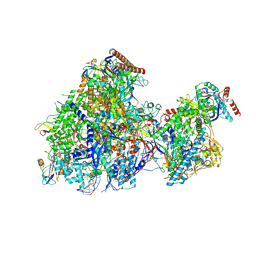 | | Cryo EM structure of Komagataella phaffii RNAPII-Rat1-Rai1 pre-termination complex | | Descriptor: | 5'-3' exoribonuclease, DNA (90-mer), DNA-directed RNA polymerase subunit, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8YFR
 
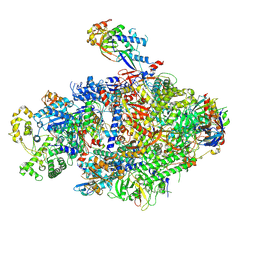 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1 complex bound within the RNAPII cleft | | Descriptor: | 5'-3' exoribonuclease, DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Murayama, Y, Yanagisawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2024-02-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8YF5
 
 | | Cryo EM structure of Komagataella phaffii Rat1-Rai1-Rtt103 complex | | Descriptor: | 5'-3' exoribonuclease, Decapping nuclease, Exonuclease Rat1p and Rai1p interacting protein | | Authors: | Yanagisawa, T, Murayama, Y, Ehara, H, Sekine, S.I. | | Deposit date: | 2024-02-24 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex.
Nat Commun, 15, 2024
|
|
8I1C
 
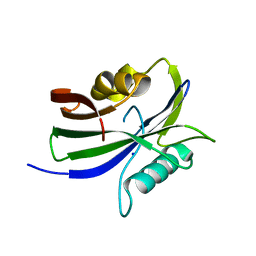 | |
8I1A
 
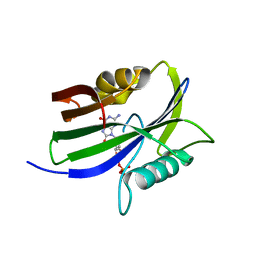 | |
8I1D
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1I
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 7.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1E
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.0 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I18
 
 | |
8I1H
 
 | |
8I1J
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP at pH 9.7 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I1G
 
 | |
8I8T
 
 | |
8I1F
 
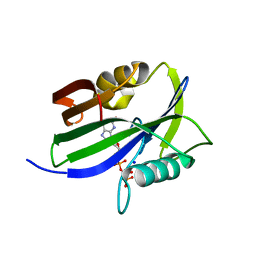 | | Crystal structure of human MTH1(G2K mutant) in complex with 2-oxo-dATP at pH 8.6 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Protonation states of Asp residues in the human Nudix hydrolase MTH1 contribute to its broad substrate recognition.
Febs Lett., 597, 2023
|
|
8I19
 
 | |
8I8S
 
 | |
8HSH
 
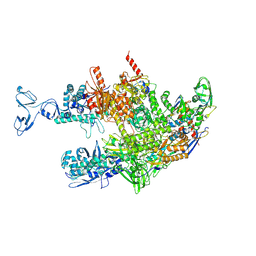 | | Thermus thermophilus RNA polymerase coreenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murayama, Y, Ehara, H, Sekine, S. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
8HSG
 
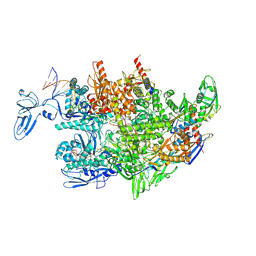 | | Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA (27-MER), DNA (37-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Murayama, Y, Ehara, H, Sekine, S. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
8HSL
 
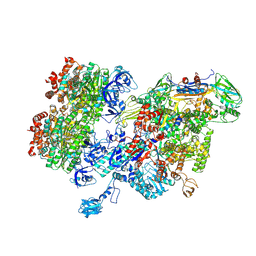 | | Thermus thermophilus RNA polymerase bound with an inverted Rho hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Murayama, Y, Ehara, H, Sekine, S. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
8HSJ
 
 | | Thermus thermophilus transcription termination factor Rho bound with ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Murayama, Y, Ehara, H, Sekine, S. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
8HSR
 
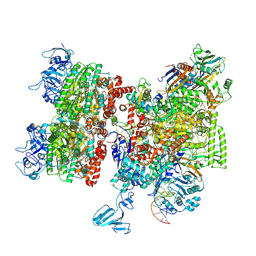 | | Thermus thermophilus Rho-engaged RNAP elongation complex (composite structure) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (31-MER), ... | | Authors: | Murayama, Y, Ehara, H, Sekine, S. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus.
Sci Adv, 9, 2023
|
|
8GZC
 
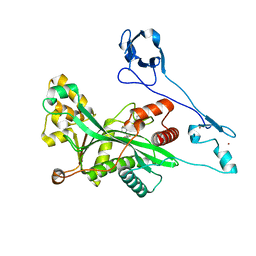 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
5ZFI
 
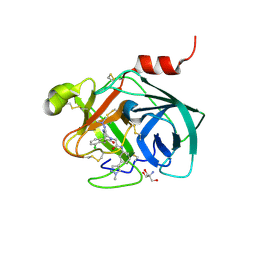 | | Mouse kallikrein 7 in complex with 6-benzyl-1,4-diazepan-7-one derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(3Z,6R)-6-[(2,6-dichlorophenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxo-1,4-diazepan-1-yl]-N-[3-(1-methyl-1H-pyrazol-4-yl)phenyl]acetamide, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
8H4V
 
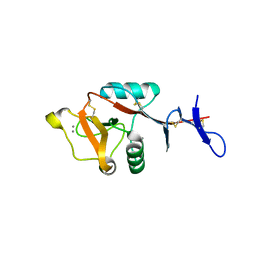 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
8HB5
 
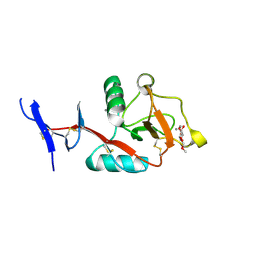 | | Crystal structure of Mincle in complex with HD-275 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
