3C4E
 
 | | Pim-1 Kinase Domain in Complex with 3-aminophenyl-7-azaindole | | Descriptor: | IMIDAZOLE, N-phenyl-1H-pyrrolo[2,3-b]pyridin-3-amine, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Zhang, K.Y.J, Wang, W. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C4F
 
 | |
1RLC
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
1RLD
 
 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5V1X
 
 | | Carbon Sulfoxide lyase, Egt2 Y134F in complex with its substrate | | Descriptor: | (1S)-2-{2-[(R)-(2R)-2-amino-2-carboxyethanesulfinyl]-1H-imidazol-4-yl}-1-carboxy-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
5UTS
 
 | |
4YGX
 
 | |
4YH1
 
 | |
4YGY
 
 | |
4ZM9
 
 | | Crystal structure of circularly permuted human asparaginase-like protein 1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CITRATE ANION, ... | | Authors: | Li, W.Z, Zhang, Y. | | Deposit date: | 2015-05-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Intramolecular Cleavage of the hASRGL1 Homodimer Occurs in Two Stages.
Biochemistry, 55, 2016
|
|
4N8N
 
 | | Crystal structure of Mycobacterial FtsX extracellular domain | | Descriptor: | Cell division protein FtsX, POTASSIUM ION | | Authors: | Mavrici, D. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N8O
 
 | |
6O6L
 
 | | The Structure of EgtB(Cabther) in complex with Hercynine | | Descriptor: | EgtB (Cabther), FE (III) ION, N,N,N-trimethyl-histidine | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
6O6M
 
 | | The Structure of EgtB (Cabther) | | Descriptor: | EgtB (Cabther), FE (III) ION, GLYCEROL | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
5TIG
 
 | |
5TT2
 
 | |
5TSU
 
 | | Active conformation for Engineered human cystathionine gamma lyase (E59N, R119L, E339V) to depleting methionine | | Descriptor: | CYSTEINE, Cystathionine gamma-lyase, METHIONINE, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2016-10-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of an Engineered Cystathionine-gamma-lyase Reveal the Critical Role of Electrostatic Interactions in the Active Site.
Biochemistry, 56, 2017
|
|
5V12
 
 | | Crystal structure of Carbon Sulfoxide lyase, Egt2 Y134F with sulfenic acid intermediate | | Descriptor: | (1S)-1-carboxy-2-[2-(hydroxysulfanyl)-1H-imidazol-4-yl]-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
6BGN
 
 | | Crystal Structure of 4-Oxalocrotonate Tautomerase After Incubation with 5-Fluoro-2-hydroxy-2,4-pentadienoate | | Descriptor: | 2-hydroxymuconate tautomerase, 5-fluoranyl-2-oxidanylidene-pentanoic acid, GLYCEROL, ... | | Authors: | Zhang, Y, Li, W, Stack, T. | | Deposit date: | 2017-10-29 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Inactivation of 4-Oxalocrotonate Tautomerase by 5-Halo-2-hydroxy-2,4-pentadienoates.
Biochemistry, 57, 2018
|
|
6OVG
 
 | |
6AKS
 
 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
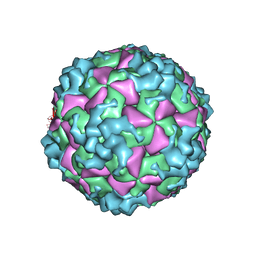 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6KW1
 
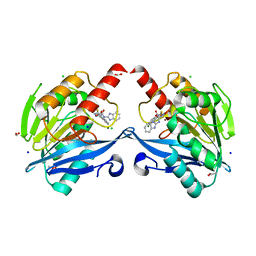 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | Descriptor: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Yang, K.W, Xiang, Y. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.775212 Å) | | Cite: | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
7VWA
 
 | |
