1PAD
 
 | |
3VU0
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VU1
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PhAglB-L, O74088_PYRHO) from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein PH0242 | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
1VID
 
 | | CATECHOL O-METHYLTRANSFERASE | | Descriptor: | 3,5-DINITROCATECHOL, CATECHOL O-METHYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Vidgren, J, Svensson, L.A, Liljas, A. | | Deposit date: | 1996-01-05 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of catechol O-methyltransferase.
Nature, 368, 1994
|
|
2W79
 
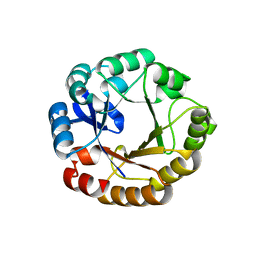 | | Establishing wild-type levels of catalytic activity on natural and artificial (ba)8-barrel protein scaffolds | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, CHLORIDE ION | | Authors: | Claren, J, Malisi, C, Hocker, B, Sterner, R. | | Deposit date: | 2008-12-21 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Establishing Wild-Type Levels of Catalytic Activity on Natural and Artificial (Beta Alpha)8- Barrel Protein Scaffolds.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1XQC
 
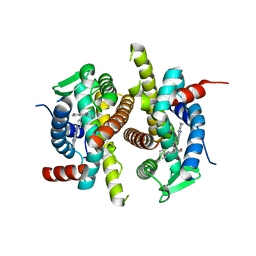 | | X-ray structure of ERalpha LBD bound to a tetrahydroisoquinoline SERM ligand at 2.05A resolution | | Descriptor: | (1S)-1-{4-[(9AR)-OCTAHYDRO-2H-PYRIDO[1,2-A]PYRAZIN-2-YL]PHENYL}-2-PHENYL-1,2,3,4-TETRAHYDROISOQUINOLIN-6-OL, Estrogen receptor | | Authors: | Renaud, J, Bischoff, S.F, Buhl, T, Floersheim, P, Fournier, B, Geiser, M, Halleux, C, Kallen, J, Keller, H.J, Ramage, P. | | Deposit date: | 2004-10-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selective Estrogen Receptor Modulators with Conformationally Restricted Side Chains. Synthesis and Structure-Activity Relationship of ERalpha-Selective Tetrahydroisoquinoline Ligands
J.Med.Chem., 48, 2005
|
|
9F9Y
 
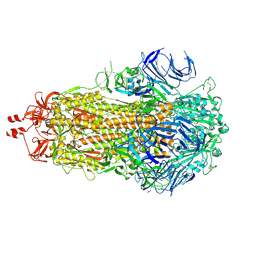 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
4R3M
 
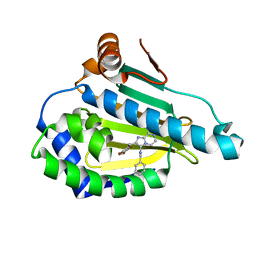 | | Crystal structure of Human Hsp90 with JR9 | | Descriptor: | Heat shock protein HSP 90-alpha, N~3~-benzyl-2-[(6-bromo-1,3-benzodioxol-5-yl)methyl]imidazo[1,2-a]pyrazine-3,8-diamine | | Authors: | Li, J, Yang, M, Ren, J, Xiong, B, He, J. | | Deposit date: | 2014-08-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multi-substituted 8-aminoimidazo[1,2-a]pyrazines by Groebke-Blackburn-Bienayme reaction and their Hsp90 inhibitory activity.
Org.Biomol.Chem., 13, 2015
|
|
1LWE
 
 | | CRYSTAL STRUCTURE OF M41L/T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMN) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LWC
 
 | | CRYSTAL STRUCTURE OF M184V MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1JLQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1TKX
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
1LW2
 
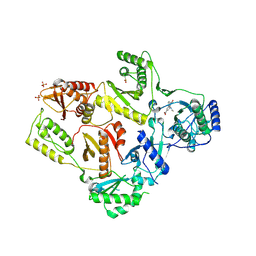 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
8QTD
 
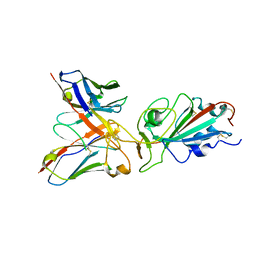 | | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | | Descriptor: | Spike glycoprotein,Fibritin, XBB-7 fab heavy chain, XBB-7 fab light chain | | Authors: | Ren, J, Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2023-10-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
4L8Z
 
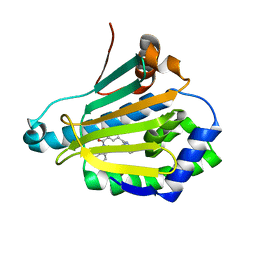 | | Crystal structure of Human Hsp90 with RL1 | | Descriptor: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](3,4-dihydroisoquinolin-2(1H)-yl)methanone | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8QSQ
 
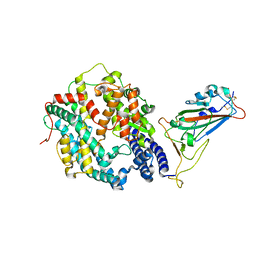 | |
4L91
 
 | | Crystal structure of Human Hsp90 with X29 | | Descriptor: | 4-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-6-chlorobenzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4L94
 
 | | Crystal structure of Human Hsp90 with S46 | | Descriptor: | (4-hydroxyphenyl)(4-methylpiperazin-1-yl)methanone, Heat shock protein HSP 90-alpha | | Authors: | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Identification of a new series of potent diphenol HSP90 inhibitors by fragment merging and structure-based optimization
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3NAR
 
 | | Crystal structure of ZHX1 HD4 (zinc-fingers and homeoboxes protein 1, homeodomain 4) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 1 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
3NAU
 
 | | Crystal structure of ZHX2 HD2 (zinc-fingers and homeoboxes protein 2, homeodomain 2) | | Descriptor: | SULFATE ION, Zinc fingers and homeoboxes protein 2 | | Authors: | Ren, J, Bird, L.E, Owens, R.J, Stammers, D.K, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel structural features in two ZHX homeodomains derived from a systematic study of single and multiple domains
Bmc Struct.Biol., 10, 2010
|
|
5BNQ
 
 | | Crystal structure of hRANKL-mRANK complex | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Ren, J. | | Deposit date: | 2015-05-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A RANKL mutant used as an inter-species vaccine for efficient immunotherapy of osteoporosis.
Sci Rep, 5, 2015
|
|
4OEW
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-iodo-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
4OEX
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
