6SBA
 
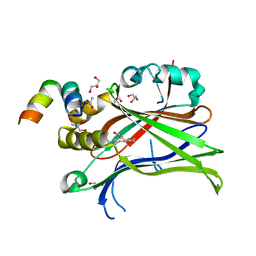 | | Crystal Structure of mTEAD with a VGL4 Tertiary Structure Mimetic | | Descriptor: | ACETYL GROUP, GLYCEROL, Transcriptional enhancer factor TEF-3, ... | | Authors: | Adihou, H, Grossmann, T.N, Waldmann, H, Gasper, R. | | Deposit date: | 2019-07-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A protein tertiary structure mimetic modulator of the Hippo signalling pathway.
Nat Commun, 11, 2020
|
|
1D6E
 
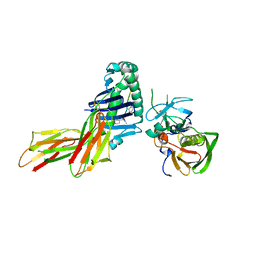 | | CRYSTAL STRUCTURE OF HLA-DR4 COMPLEX WITH PEPTIDOMIMETIC AND SEB | | Descriptor: | ENTEROTOXIN TYPE B, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, PEPTIDOMIMETIC INHIBITOR | | Authors: | Swain, A, Crowther, R, Kammlott, U. | | Deposit date: | 1999-10-13 | | Release date: | 2000-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Peptide and peptide mimetic inhibitors of antigen presentation by HLA-DR class II MHC molecules. Design, structure-activity relationships, and X-ray crystal structures.
J.Med.Chem., 43, 2000
|
|
6DUY
 
 | |
6DUW
 
 | |
6DV1
 
 | |
6E4F
 
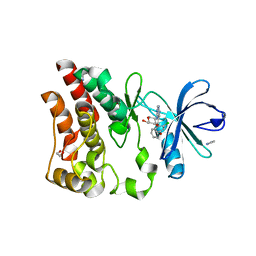 | | Crystal structure of ARQ 531 in complex with the kinase domain of BTK | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-2-{[5-(2-chloro-4-phenoxybenzene-1-carbonyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2,3,4-trideoxy-D-erythro-hexitol, IMIDAZOLE, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The BTK Inhibitor ARQ 531 Targets Ibrutinib-Resistant CLL and Richter Transformation.
Cancer Discov, 8, 2018
|
|
4EJN
 
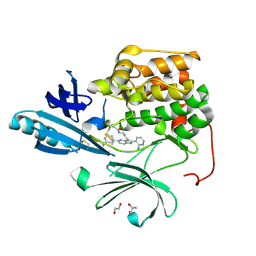 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
6CFD
 
 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
4TVM
 
 | |
4TVO
 
 | |
3C7X
 
 | | Hemopexin-like domain of matrix metalloproteinase 14 | | Descriptor: | CHLORIDE ION, Matrix metalloproteinase-14, SODIUM ION | | Authors: | Tochowicz, A, Itoh, Y, Maskos, K, Bode, W, Goettig, P. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions
J.Biol.Chem., 286, 2011
|
|
5NZ9
 
 | | NMR structure of an EphA2-Sam fragment | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Sam-Sam interaction between Ship2 and the EphA2 receptor: design and analysis of peptide inhibitors.
Sci Rep, 7, 2017
|
|
6O2W
 
 | |
6O2U
 
 | | Crystal structure of the SARAF luminal domain | | Descriptor: | Store-operated calcium entry-associated regulatory factor | | Authors: | Kimberlin, C.R, Minor, D.L. | | Deposit date: | 2019-02-24 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARAF Luminal Domain Structure Reveals a Novel Domain-Swapped beta-Sandwich Fold Important for SOCE Modulation.
J.Mol.Biol., 431, 2019
|
|
6O2V
 
 | |
6PE6
 
 | | PANK3 complex structure with compound PZ-3022 | | Descriptor: | 6-{4-[(4-cyclopropylphenyl)acetyl]piperazin-1-yl}pyridazine-3-carbonitrile, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pantothenate kinase activation relieves coenzyme A sequestration and improves mitochondrial function in mice with propionic acidemia.
Sci Transl Med, 13, 2021
|
|
3UDL
 
 | | 3-heterocyclyl quinolone bound to HCV NS5B | | Descriptor: | 3-(5-benzyl-1,2,4-oxadiazol-3-yl)-6-fluoro-1-[2-fluoro-4-(trifluoromethyl)benzyl]-7-(4-methylpiperazin-1-yl)quinolin-4(1H)-one, HCV NS5B polymerase | | Authors: | Somoza, J.R. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Quinolones as HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6X2W
 
 | | Crystal Structure of PKINES peptide bound to CRM1(E571K) | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2U
 
 | | Crystal Structure of PKINES peptide bound to CRM1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2S
 
 | | Crystal Structure of Mek1(NQ)NES peptide bound to CRM | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2M
 
 | | Crystal Structure of unliganded CRM1-Ran-RanBP1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2P
 
 | | Crystal Structure of the Mek1NES peptide bound to CRM1 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Exportin-1, GLYCEROL, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
6X2V
 
 | | Crystal Structure of PKI(DE)NES peptide bound to CRM1 | | Descriptor: | Exportin-1, GLYCEROL, GTP-binding nuclear protein Ran, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
4OS7
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK607 (bicyclic) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS5
 
 | |
