3KPP
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, EEYLQAFTY, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPO
 
 | | Crystal Structure of HLA B*4403 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPM
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3MV8
 
 | | Crystal Structure of the TK3-Gln55His TCR in complex with HLA-B*3501/HPVG | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chen, Z, Miles, J.J, Liu, Y.C, Bell, M.J, Sullivan, L.C, Kjer-Nielsen, L, Brennan, R.M, Burrows, J.M, Neller, M.A, Khanna, R, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allelic polymorphism in the T cell receptor and its impact on immune responses
J.Exp.Med., 207, 2010
|
|
3MV9
 
 | | Crystal Structure of the TK3-Gln55Ala TCR in complex with HLA-B*3501/HPVG | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chen, Z, Miles, J.J, Liu, Y.C, Bell, M.J, Sullivan, L.C, Kjer-Nielsen, L, Brennan, R.M, Burrows, J.M, Neller, M.A, Khanna, R, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allelic polymorphism in the T cell receptor and its impact on immune responses
J.Exp.Med., 207, 2010
|
|
3MV7
 
 | | Crystal Structure of the TK3 TCR in complex with HLA-B*3501/HPVG | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chen, Z, Miles, J.J, Liu, Y.C, Bell, M.J, Sullivan, L.C, Kjer-Nielsen, L, Brennan, R.M, Burrows, J.M, Neller, M.A, Khanna, R, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allelic polymorphism in the T cell receptor and its impact on immune responses
J.Exp.Med., 207, 2010
|
|
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
1T0M
 
 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
5LDT
 
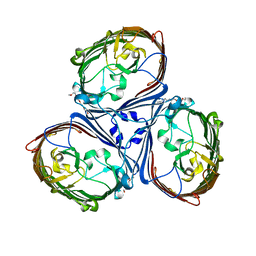 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | Authors: | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
5LDV
 
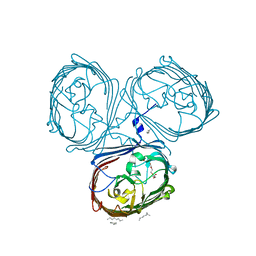 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | CALCIUM ION, MOMP porin, N-OCTANE, ... | | Authors: | Wallat, G.D, Ferrara, L.M.G, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
1G2N
 
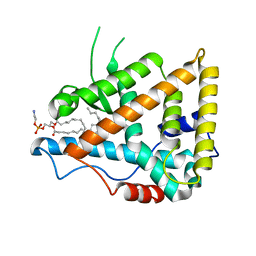 | | CRYSTAL STRUCTURE OF THE LIGAND BINDING DOMAIN OF THE ULTRASPIRACLE PROTEIN USP, THE ORTHOLOG OF RXRS IN INSECTS | | Descriptor: | L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ULTRASPIRACLE PROTEIN | | Authors: | Billas, I.M.L, Moulinier, L, Rochel, N, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-10-20 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects.
J.Biol.Chem., 276, 2001
|
|
2CJW
 
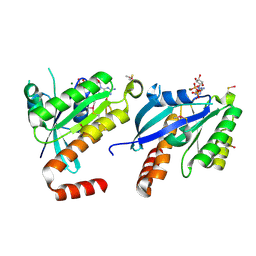 | | Crystal structure of the small GTPase Gem (GemDNDCaM) in complex to Mg.GDP | | Descriptor: | GTP-BINDING PROTEIN GEM, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Splingard, A, Menetrey, J, Perderiset, M, Cicolari, J, Hamoudi, F, Cabanie, L, El Marjou, A, Wells, A, Houdusse, A, de Gunzburg, J. | | Deposit date: | 2006-04-09 | | Release date: | 2006-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Characterization of the Gem Gtpase.
J.Biol.Chem., 282, 2007
|
|
3NEL
 
 | | Aspartyl-tRNA synthetase complexed with aspartic acid | | Descriptor: | ASPARTIC ACID, Aspartyl-tRNA synthetase | | Authors: | Schmitt, E, Moras, D, Moulinier, L. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation
Embo J., 17, 1998
|
|
3NEM
 
 | | Aspartyl-tRNA synthetase complexed with aspartyl adenylate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, Aspartyl-tRNA synthetase, ... | | Authors: | Schmitt, E, Moras, D, Moulinier, L. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation
Embo J., 17, 1998
|
|
3NEN
 
 | |
2L7S
 
 | |
6Y47
 
 | |
6Z33
 
 | |
6Z2N
 
 | |
6YY5
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with TCV_L5 | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[2-[[(2~{S})-3-[[(2~{S})-3-[[1-[2-[2-[2-[4-[4-[5-(acetamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]-2-fluoranyl-phenyl]piperazin-1-yl]-2-oxidanylidene-ethoxy]ethoxy]ethyl]-1,2,3-triazol-4-yl]methylamino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-oxidanylidene-ethyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Naismith, J.H, Moynie, L.M. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa.
Acs Infect Dis., 2022
|
|
4YF5
 
 | |
4YF6
 
 | | Crystal structure of oxidised Rv1284 | | Descriptor: | Beta-carbonic anhydrase 1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Hofmann, A. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Chemical probing suggests redox-regulation of the carbonic anhydrase activity of mycobacterial Rv1284.
Febs J., 282, 2015
|
|
4YF4
 
 | |
2XUE
 
 | | CRYSTAL STRUCTURE OF JMJD3 | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Rowland, P, Mosley, J, Thomas, P.J. | | Deposit date: | 2010-10-19 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
4ASK
 
 | | CRYSTAL STRUCTURE OF JMJD3 WITH GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, COBALT (II) ION, LYSINE-SPECIFIC DEMETHYLASE 6B, ... | | Authors: | Chung, C, Mosley, J, Liddle, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response
Nature, 488, 2012
|
|
