5GL3
 
 | | Crystal structure of TON_0340 in complex with Mg | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GL4
 
 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
5GKX
 
 | | Crystal structure of TON_0340, apo form | | Descriptor: | PHOSPHATE ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
8JOQ
 
 | |
8JOY
 
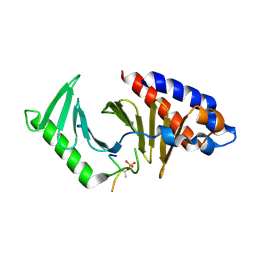 | | Plk1 polo-box domain bound to HPV4 L2 residues 251-257 with pThr255 | | Descriptor: | Peptide from Minor capsid protein L2, Serine/threonine-protein kinase PLK1 | | Authors: | Ku, B, Jung, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Plk1 Polo-Box Domain Bound to the Human Papillomavirus Minor Capsid Protein L2-Derived Peptide.
J.Microbiol, 61, 2023
|
|
8J9Q
 
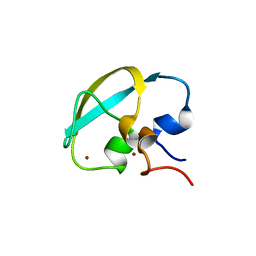 | | Crystal structure of UBR box of UBR4 apo | | Descriptor: | E3 ubiquitin-protein ligase UBR4, ZINC ION | | Authors: | Jeong, D.-E, KIm, S.-J, Shin, H.-C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates.
Commun Biol, 6, 2023
|
|
8J9R
 
 | |
1RYQ
 
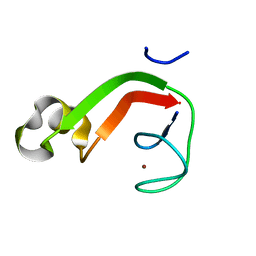 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7CIN
 
 | |
8CB1
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-PNT-DNM 15 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[5-(phenanthren-9-ylmethoxy)pentyl]piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
8CB6
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in covalent complex with TAMRA tagged 1,6-Epi-cylcophellitol aziridine activity based probe | | Descriptor: | (1S,2R,3R,4R,5R)-5-[8-[4-(4-azanylbutyl)-1,2,3-triazol-1-yl]octylamino]-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
3TDK
 
 | |
2P6Z
 
 | |
2P7S
 
 | |
6JM4
 
 | |
6L6R
 
 | | Crystal structure of LRP6 E1E2-SOST complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Choi, H.-J, Kim, J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains.
Nat Commun, 11, 2020
|
|
5XC5
 
 | | Crystal structure of Acanthamoeba polyphaga mimivirus Rab GTPase in complex with GTP | | Descriptor: | ACETATE ION, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ku, B, You, J.A, Kim, S.J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystal structures of two forms of the Acanthamoeba polyphaga mimivirus Rab GTPase
Arch. Virol., 162, 2017
|
|
5GOT
 
 | | Crystal structure of SP-PTP, low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, Low molecular weight phosphotyrosine phosphatase family protein | | Authors: | Ku, B, Keum, C.W, KIim, S.J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of SP-PTP, a low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes
Biochem.Biophys.Res.Commun., 478, 2016
|
|
5YZH
 
 | | Crystal Structure of Mouse Cytosolic Isocitrate Dehydrogenase | | Descriptor: | GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | NADP+-dependent cytosolic isocitrate dehydrogenase provides NADPH in the presence of cadmium due to the moderate chelating effect of glutathione.
J. Biol. Inorg. Chem., 23, 2018
|
|
5YZI
 
 | | Crystal Structure of Mouse Cytosolic Isocitrate Dehydrogenase complexed with Cadmium | | Descriptor: | CADMIUM ION, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cho, H.J, Kang, B.S. | | Deposit date: | 2017-12-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | NADP+-dependent cytosolic isocitrate dehydrogenase provides NADPH in the presence of cadmium due to the moderate chelating effect of glutathione.
J. Biol. Inorg. Chem., 23, 2018
|
|
5XC3
 
 | |
2JG7
 
 | | Crystal structure of Seabream Antiquitin and Elucidation of its substrate specificity | | Descriptor: | ANTIQUITIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tang, W.K, Wong, K.B, Cha, S.S, Lee, H.S, Cheng, C.H.K, Fong, W.P. | | Deposit date: | 2007-02-09 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Seabream Antiquitin Reveals the Structural Basis of its Substrate Specificity.
FEBS Lett., 582, 2008
|
|
5I23
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF022 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-{[(1S,2R,3S,4S,5R,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)cyclohexyl]amino}octyl)triaza-1,2-dien-2-ium, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
8B0D
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, ALANINE, SULFATE ION, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8B0B
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
