8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
3OSY
 
 | |
3SO6
 
 | |
3THB
 
 | | Structure of PLK1 kinase domain in complex with a benzolactam-derived inhibitor | | Descriptor: | 9-chloro-2-({5-[3-(dimethylamino)propyl]-2-methylpyridin-3-yl}amino)-5,7-dihydro-6H-pyrimido[5,4-d][1]benzazepine-6-thi one, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-08-18 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Benzolactam-Derived Inhibitor of Polo-Like Kinase 1 (MLN0905).
J.Med.Chem., 55, 2012
|
|
4ZXD
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD | | Descriptor: | Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
4ZXC
 
 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Fe3+ | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
4ZXA
 
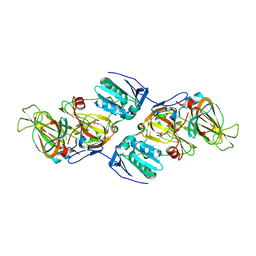 | | Crystal Structure of hydroquinone 1,2-dioxygenase PnpCD in complex with Cd2+ and 4-hydroxybenzonitrile | | Descriptor: | 4-hydroxybenzonitrile, CADMIUM ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Liu, S, Su, T, Zhang, C, Gu, L. | | Deposit date: | 2015-05-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of PnpCD, a Two-subunit Hydroquinone 1,2-Dioxygenase, Reveals a Novel Structural Class of Fe2+-dependent Dioxygenases.
J.Biol.Chem., 290, 2015
|
|
3HU1
 
 | | Structure of p97 N-D1 R95G mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3HU2
 
 | | Structure of p97 N-D1 R86A mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
3HU3
 
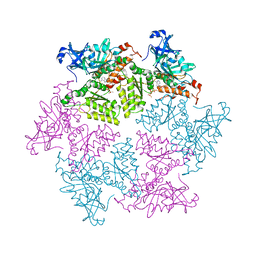 | | Structure of p97 N-D1 R155H mutant in complex with ATPgS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Tang, W.-K. | | Deposit date: | 2009-06-12 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants.
Embo J., 29, 2010
|
|
4ES4
 
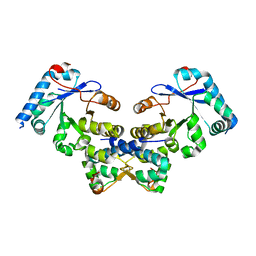 | | Crystal structure of YdiV and FlhD complex | | Descriptor: | Flagellar transcriptional regulator FlhD, Putative cyclic di-GMP regulator CdgR | | Authors: | Li, B, Gu, L. | | Deposit date: | 2012-04-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility
Nucleic Acids Res., 40, 2012
|
|
5GWN
 
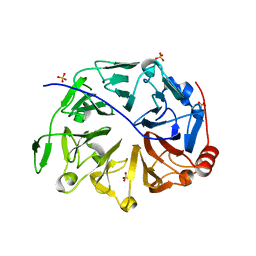 | | Crystal structure of human RCC2 | | Descriptor: | Protein RCC2, SULFATE ION | | Authors: | Liang, L, Yun, C.H, Yin, Y.X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | RCC2 is a novel p53 target in suppressing metastasis.
Oncogene, 37, 2018
|
|
7XS2
 
 | | Monomer structure of HtrA from Helicobacter pylori | | Descriptor: | Periplasmic serine endoprotease DegP-like | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
7XS0
 
 | | Trimer structure of HtrA from Helicobacter pylori bound with a tripeptide | | Descriptor: | Periplasmic serine endoprotease DegP-like, UNK-UNK-UNK | | Authors: | Cui, L, Liu, W. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures and solution conformations of HtrA from Helicobacter pylori reveal pH-dependent oligomeric conversion and conformational rearrangements.
Int.J.Biol.Macromol., 243, 2023
|
|
5GMZ
 
 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | Descriptor: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
4HVB
 
 | | Catalytic unit of PI3Kg in complex with PI3K/mTOR dual inhibitor PF-04979064 | | Descriptor: | 1-{1-[(2S)-2-hydroxypropanoyl]piperidin-4-yl}-3-methyl-8-(6-methylpyridin-3-yl)-1,3-dihydro-2H-imidazo[4,5-c][1,5]naphthyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knighton, D.R, Cheng, H. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Highly Potent PI3K/mTOR Dual Inhibitor PF-04979064 through Structure-Based Drug Design.
ACS Med Chem Lett, 4, 2013
|
|
7YDQ
 
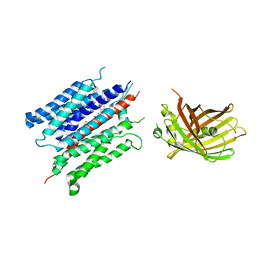 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | Descriptor: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | Authors: | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
4ZAE
 
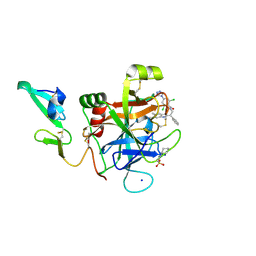 | | Development of a novel class of potent and selective FIXa inhibitors | | Descriptor: | 2,6-dichloro-N-[(2R)-2-(5,6-dimethyl-1H-benzimidazol-2-yl)-2-phenylethyl]-4-(4H-1,2,4-triazol-4-yl)benzamide, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, ... | | Authors: | Hruza, A, Reichert, P. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a novel class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
