2DJY
 
 | | Solution structure of Smurf2 WW3 domain-Smad7 PY peptide complex | | Descriptor: | Mothers against decapentaplegic homolog 7, Smad ubiquitination regulatory factor 2 | | Authors: | Chong, P.A, Lin, H, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2006-04-06 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Expanded WW Domain Recognition Motif Revealed by the Interaction between Smad7 and the E3 Ubiquitin Ligase Smurf2.
J.Biol.Chem., 281, 2006
|
|
6MD4
 
 | |
6MD1
 
 | |
5CEO
 
 | | DLK in complex with inhibitor 2-((6-(3,3-difluoropyrrolidin-1-yl)-4-(1-(oxetan-3-yl)piperidin-4-yl)pyridin-2-yl)amino)isonicotinonitrile | | Descriptor: | 2-[[6-[3,3-bis(fluoranyl)pyrrolidin-1-yl]-4-[1-(oxetan-3-yl)piperidin-4-yl]pyridin-2-yl]amino]pyridine-4-carbonitrile, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5CEN
 
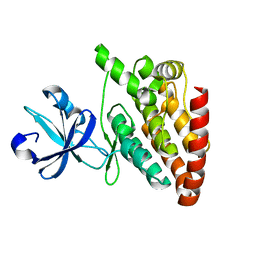 | | Crystal structure of DLK (kinase domain) | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5CEP
 
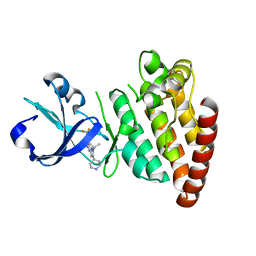 | |
5CEQ
 
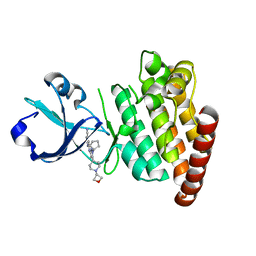 | |
8T4C
 
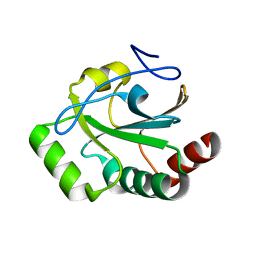 | |
6UK1
 
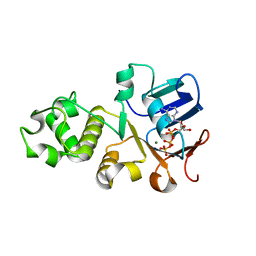 | | Crystal structure of nucleotide-binding domain 2 (NBD2) of the human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Vorobiev, S.M, Vernon, R.M, Khazanov, N, Senderowitz, H, Forman-Kay, J.D, Hunt, J.F. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | A thermodynamically stabilized form of the second nucleotide binding domain from human CFTR shows a catalytically inactive conformation
To Be Published
|
|
6WED
 
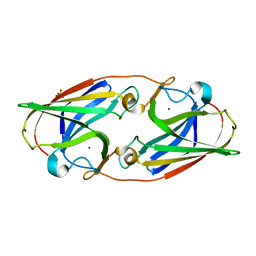 | |
6WEE
 
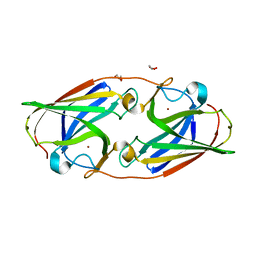 | | Copper-bound M88I variant of Campylobacter jejuni P19 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Chan, A.C, Murphy, M.E. | | Deposit date: | 2020-04-02 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A copper site is required for iron transport by the periplasmic proteins P19 and FetP.
Metallomics, 12, 2020
|
|
6WEF
 
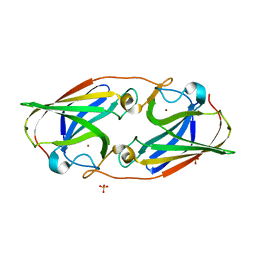 | |
3RIY
 
 | |
3RIG
 
 | |
3ZG6
 
 | | The novel de-long chain fatty acid function of human sirt6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-6, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2012-12-15 | | Release date: | 2013-04-03 | | Last modified: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sirt6 Regulates Tnf-Alpha Secretion Through Hydrolysis of Long-Chain Fatty Acyl Lysine
Nature, 496, 2013
|
|
6AVI
 
 | |
6AUG
 
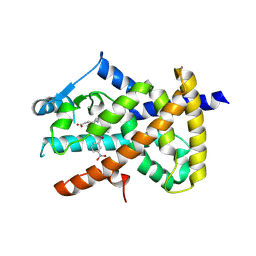 | |
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
5UGM
 
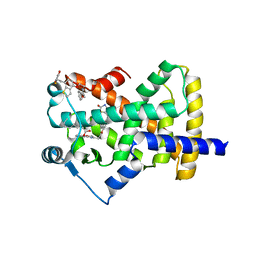 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
5UV1
 
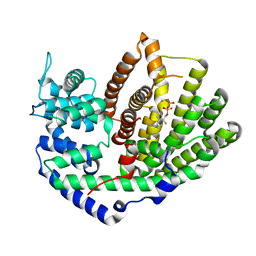 | | Crystal Structure of (+)-Limonene Synthase Complexed with 2-Fluorogeranyl Diphosphate | | Descriptor: | (+)-limonene synthase, (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, MANGANESE (II) ION | | Authors: | Prem Kumar, R, Malik, K, Oprian, D.D. | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Early Michaelis Complexes in the Reaction Catalyzed by (+)-Limonene Synthase from Citrus sinensis Using Fluorinated Substrate Analogues.
Biochemistry, 56, 2017
|
|
5UV2
 
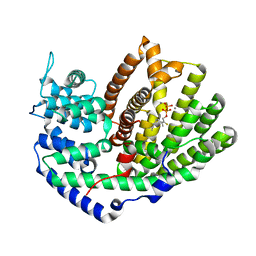 | | Crystal Structure of (+)-Limonene Synthase Complexed with 2-Fluoroneryl Diphosphate | | Descriptor: | (+)-limonene synthase, (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, MANGANESE (II) ION | | Authors: | Prem Kumar, R, Malik, K, Oprian, D.D. | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Early Michaelis Complexes in the Reaction Catalyzed by (+)-Limonene Synthase from Citrus sinensis Using Fluorinated Substrate Analogues.
Biochemistry, 56, 2017
|
|
6MCZ
 
 | |
6MD2
 
 | |
6MD0
 
 | |
6NR0
 
 | |
