8TTF
 
 | | NorA double mutant - E222QD307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
8TNW
 
 | |
8T42
 
 | | Model of TTLL6 MTBH1-2 bound to microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
8TOI
 
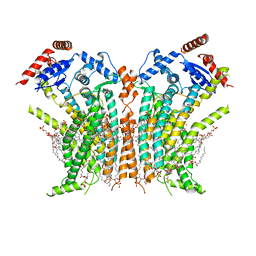 | |
5MJ6
 
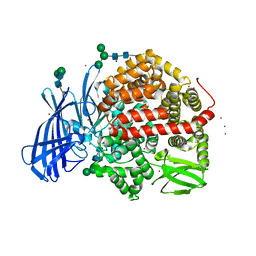 | | Ligand-induced conformational change of Insulin-regulated aminopeptidase: insights on catalytic mechanism and active site plasticity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Mpakali, A, Stratikos, E, Saridakis, E, Giastas, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Ligand-Induced Conformational Change of Insulin-Regulated Aminopeptidase: Insights on Catalytic Mechanism and Active Site Plasticity.
J. Med. Chem., 60, 2017
|
|
8TOL
 
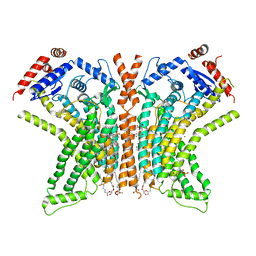 | |
1B5S
 
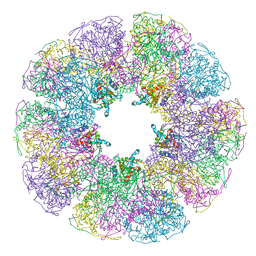 | | DIHYDROLIPOYL TRANSACETYLASE (E.C.2.3.1.12) CATALYTIC DOMAIN (RESIDUES 184-425) FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Izard, T, Aevarsson, A, Allen, M.D, Westphal, A.H, Perham, R.N, De Kok, A, Hol, W.G. | | Deposit date: | 1999-01-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1IRI
 
 | | Crystal structure of human autocrine motility factor complexed with an inhibitor | | Descriptor: | ERYTHOSE-4-PHOSPHATE, autocrine motility factor | | Authors: | Tanaka, N, Haga, A, Uemura, H, Akiyama, H, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2001-10-08 | | Release date: | 2002-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies.
J.Mol.Biol., 318, 2002
|
|
4C2D
 
 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
8Q3U
 
 | | Crystal structure of a fentanyl derivative in complex with human CA VII | | Descriptor: | Carbonic anhydrase 7, GLYCEROL, ZINC ION, ... | | Authors: | Alterio, V, Di Fiore, A, De Simone, G. | | Deposit date: | 2023-08-04 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of a novel series of potent carbonic anhydrase inhibitors with selective affinity for mu Opioid receptor for Safer and long-lasting analgesia.
Eur.J.Med.Chem., 260, 2023
|
|
8TPP
 
 | |
8TTE
 
 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
4C2E
 
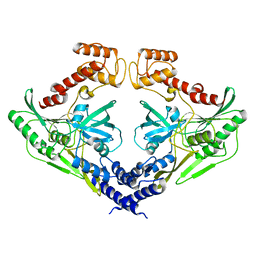 | | Crystal structure of the protease CtpB(S309A) present in a resting state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
8TPT
 
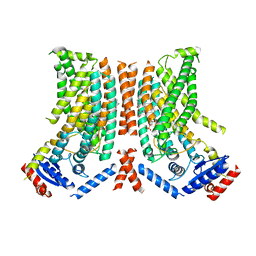 | |
4C59
 
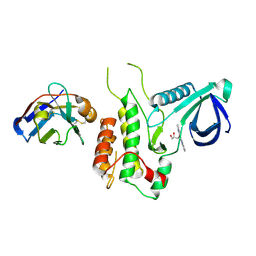 | | Structure of GAK kinase in complex with nanobody (NbGAK_4) | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cyclin-G-associated kinase, NANOBODY | | Authors: | Chaikuad, A, Keates, T, Allerston, C.K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Muller-Knapp, S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies.
Biochem. J., 459, 2014
|
|
5MBW
 
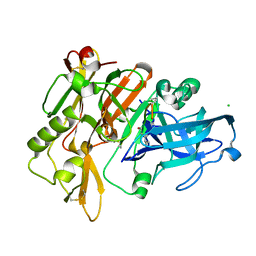 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH Pep#3 | | Descriptor: | BACE1 INHIBITOR PEPTIDE Pep#3, Beta-secretase 1, CHLORIDE ION | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
8TNX
 
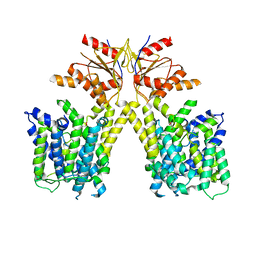 | |
8TTG
 
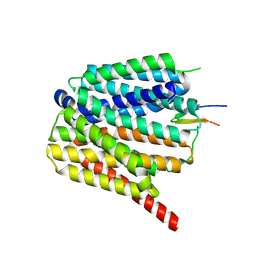 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
5MCO
 
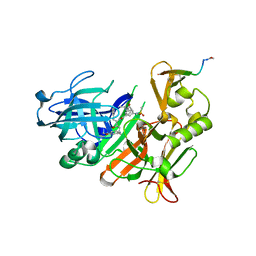 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
8TNY
 
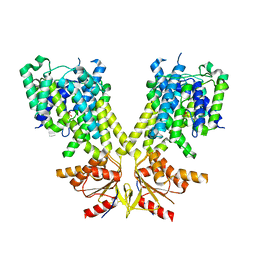 | |
1J7W
 
 | | Crystal structure of deoxy HbbetaYQ, a site directed mutant of HbA | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
4BNG
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-pentyl- 2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
1IVY
 
 | | PHYSIOLOGICAL DIMER HPP PRECURSOR | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudenko, G, Bonten, E, D'Azzo, A, Hol, W.G.J. | | Deposit date: | 1996-06-12 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of the human 'protective protein': structure of the precursor form suggests a complex activation mechanism.
Structure, 3, 1995
|
|
8TG5
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with branched 2'-PO4 RNA | | Descriptor: | (2S,3R,4R,5S)-2-(6-amino-9H-purin-9-yl)-4-{[(S)-{[(2R,3S,4R,5S)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-({[(S)-{[(2R,3R,4S,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)oxolan-3-yl dihydrogen phosphate (non-preferred name), CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
