7E2Q
 
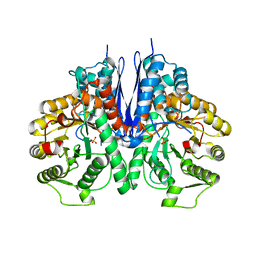 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
8EKY
 
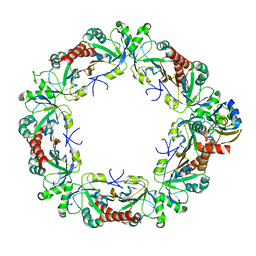 | | Cryo-EM structure of the human PRDX4-ErP46 complex | | Descriptor: | Peroxiredoxin-4, Thioredoxin domain-containing protein 5 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EKW
 
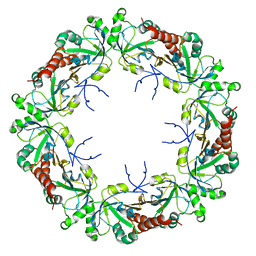 | | Cryo-EM structure of human PRDX4 | | Descriptor: | Peroxiredoxin-4 | | Authors: | Su, C.C. | | Deposit date: | 2022-09-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
8EM2
 
 | |
3IKF
 
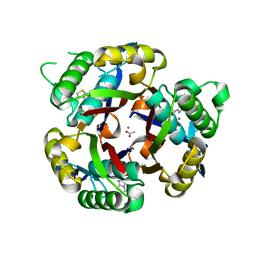 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
8FXF
 
 | | Crystal structure of the coiled-coil domain of TRIM56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase TRIM56 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TRIM56 coiled-coil domain structure provides insights into its E3 ligase functions.
Comput Struct Biotechnol J, 21, 2023
|
|
3JVH
 
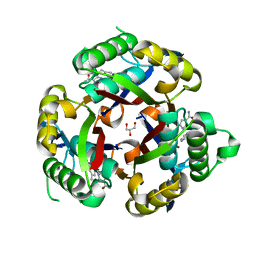 | |
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
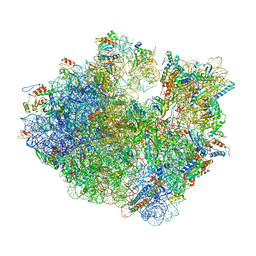 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G38
 
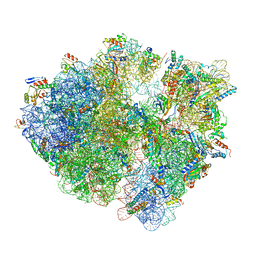 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
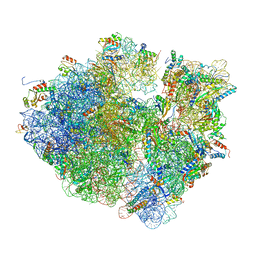 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
3L72
 
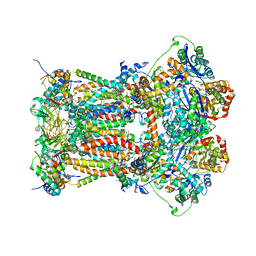 | | Chicken cytochrome BC1 complex with kresoxim-I-dimethyl bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME B, ... | | Authors: | Huang, L, Zhang, Z, Berry, E.A. | | Deposit date: | 2009-12-27 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Famoxadone and related inhibitors bind like methoxy acrylate inhibitors in the Qo site of the BC1 compl and fix the rieske iron-sulfur protein in a positio close to but distinct from that seen with stigmatellin and other "DISTAL" Qo inhibitors.
To be Published
|
|
7XLB
 
 | |
6IZQ
 
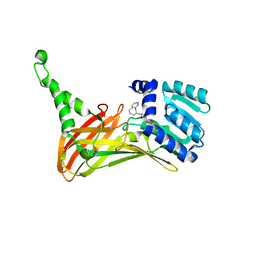 | | PRMT4 bound with a bicyclic compound | | Descriptor: | (2R)-1-(methylamino)-3-(1,3,4,5-tetrahydro-2-benzazepin-2-yl)propan-2-ol, Histone-arginine methyltransferase CARM1 | | Authors: | Xiong, B, Cao, D.Y, Guo, Z.H, Li, Y.L, Li, J, Huang, X, Shen, J.K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Design and Synthesis of Potent, Selective Inhibitors of Protein Arginine Methyltransferase 4 against Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
6VFE
 
 | | Gasdermin D pore | | Descriptor: | Gasdermin-D, N-terminal | | Authors: | Xia, S, Ruan, J, Wu, H. | | Deposit date: | 2020-01-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Gasdermin D pore structure reveals preferential release of mature interleukin-1.
Nature, 593, 2021
|
|
6UKA
 
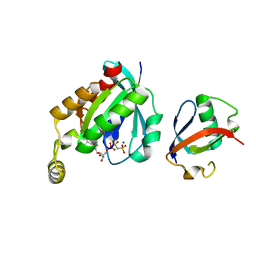 | | Crystal structure of RHOG and ELMO complex | | Descriptor: | Engulfment and cell motility protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jo, C.H, Killoran, R.C, Smith, M.J. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
5USF
 
 | |
6WIM
 
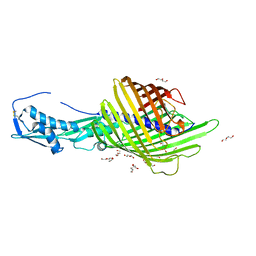 | | CdiB from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Outer membrane transporter CdiB | | Authors: | Guerin, J, Botos, I, Buchanan, S.K. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into toxin secretion by contact dependent growth inhibition transporters.
Elife, 9, 2020
|
|
5V49
 
 | |
5VQM
 
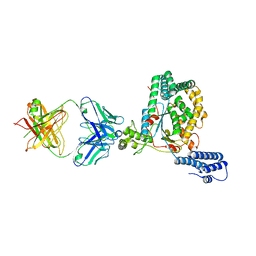 | | Clostridium difficile TcdB-GTD bound to PA41 Fab | | Descriptor: | PA41 FAB HEAVY CHAIN, PA41 FAB LIGHT CHAIN, Toxin B | | Authors: | Kroh, H.K, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2017-05-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A neutralizing antibody that blocks delivery of the enzymatic cargo of Clostridium difficile toxin TcdB into host cells.
J. Biol. Chem., 293, 2018
|
|
5VMT
 
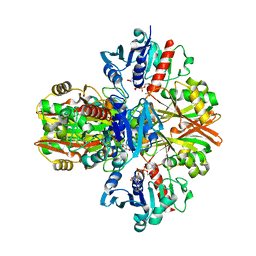 | |
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
