2XKF
 
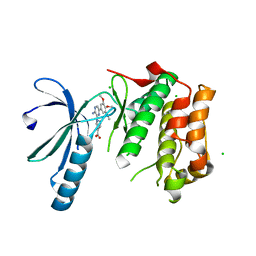 | | Structure of Nek2 bound to aminopyrazine compound 2 | | Descriptor: | 1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XNM
 
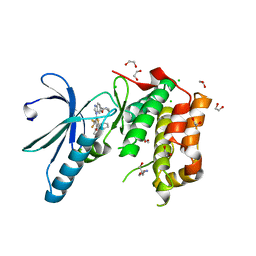 | | Structure of NEK2 bound to CCT | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-{6-[(1-METHYLPIPERIDIN-4-YL)OXY]-1H-BENZIMIDAZOL-1-YL}-3-{(1R)-1-[2-(TRIFLUOROMETHYL)PHENYL]ETHOXY}THIOPHENE-2-CARBOXAMIDE, ... | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Benzimidazole Inhibitors Induce a Dfg-Out Conformation of Never in Mitosis Gene A-Related Kinase 2 (Nek2) without Binding to the Back Pocket and Reveal a Nonlinear Structure-Activity Relationship.
J.Med.Chem., 54, 2011
|
|
1WW2
 
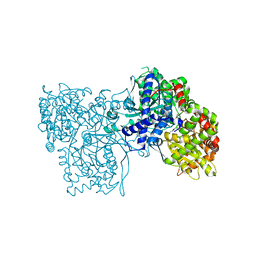 | | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-D-glucopyranosylamine and N-trifluoroacetyl-beta-D-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Anagnostou, E, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Hadjiloi, T, Tiraidis, C, Zographos, S.E, Gyorgydeak, Z, Somsak, L, Docsa, T, Gergely, P, Kolisis, F.N, Oikonomakos, N.G. | | Deposit date: | 2004-12-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-d-glucopyranosylamine and N-trifluoroacetyl-beta-d-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase
Bioorg.Med.Chem., 14, 2006
|
|
6JIU
 
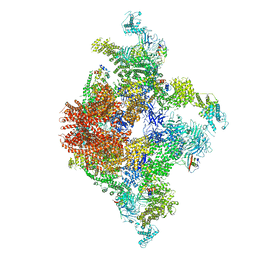 | | Structure of RyR2 (F/A/C/L-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
1WW3
 
 | | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-D-glucopyranosylamine and N-trifluoroacetyl-beta-D-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase | | Descriptor: | Glycogen phosphorylase, muscle form, N-(trifluoroacetyl)-beta-D-glucopyranosylamine, ... | | Authors: | Anagnostou, E, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Hadjiloi, T, Tiraidis, C, Zographos, S.E, Gyorgydeak, Z, Somsak, L, Docsa, T, Gergely, P, Kolisis, F.N, Oikonomakos, N.G. | | Deposit date: | 2004-12-31 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies on two bioisosteric analogues, N-acetyl-beta-d-glucopyranosylamine and N-trifluoroacetyl-beta-d-glucopyranosylamine, potent inhibitors of muscle glycogen phosphorylase
Bioorg.Med.Chem., 14, 2006
|
|
6JI8
 
 | | Structure of RyR2 (F/apoCaM dataset) | | Descriptor: | Calmodulin-1, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-20 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JI0
 
 | | Structure of RyR2 (F/A/C/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JIY
 
 | | Structure of RyR2 (F/A/C/H-Ca2+/Ca2+CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-24 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JGZ
 
 | | Structure of RyR2 (F/P/Ca2+ dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JG3
 
 | | Cryo-EM structure of RyR2 (Ca2+ alone dataset) | | Descriptor: | Ryanodine receptor 2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JRS
 
 | | Structure of RyR2 (*F/A/C/L-Ca2+/Ca2+-CaM dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JV2
 
 | | Structure of RyR2 (P/L-Ca2+/Ca2+-CaM dataset) | | Descriptor: | CALCIUM ION, Calmodulin-1, RyR2, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JHN
 
 | | Structure of RyR2 (F/C/Ca2+ dataset) | | Descriptor: | CAFFEINE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JII
 
 | | Structure of RyR2 (F/A/C/L-Ca2+/apo-CaM-M dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JRR
 
 | | Structure of RyR2 (*F/A/C/L-Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
6JFW
 
 | | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket | | Descriptor: | PA0833-PD protein | | Authors: | Lin, X, Ye, F, Lin, S, Yang, F.L, Chen, Z.M, Cao, Y, Chen, Z.J, Gu, J, Lu, G.W. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of PA0833 periplasmic domain from Pseudomonas aeruginosa reveals an unexpected enlarged peptidoglycan binding pocket.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6JH6
 
 | | Structure of RyR2 (F/A/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | Descriptor: | Glycoprotein, scFv 523-11 | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9037 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | Descriptor: | Glycoprotein,Glycoprotein,Glycoprotein | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
2AMV
 
 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
7C2J
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (without additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
7AY9
 
 | | Crystal structure of CK2 bound by compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-(5-(6-oxo-1,6-dihydropyridin-3-yl)-1-(2-(piperidin-1-yl)ethyl)-1H-1,2,3-triazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metadynamics simulations of CK2 compound unbinding to understand slow dissociation kinetics.
To Be Published
|
|
7AYA
 
 | | Crystal structure of CK2 bound by compound 9 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Metadynamics simulations of CK2 compound unbinding to understand slow dissociation kinetics.
To Be Published
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
1OMG
 
 | | NMR STUDY OF OMEGA-CONOTOXIN MVIIA | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Kohno, T, Kim, J.-I, Kobayashi, K, Kodera, Y, Maeda, T, Sato, K. | | Deposit date: | 1995-04-26 | | Release date: | 1996-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the calcium channel blocker omega-conotoxin MVIIA.
Biochemistry, 34, 1995
|
|
