2JLX
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA and ADP-Vanadate | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FI9
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI6
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI5
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | GLYCEROL, MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI8
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
7FI7
 
 | | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D | | Descriptor: | MHC class I polypeptide-related sequence A, NKG2-D type II integral membrane protein | | Authors: | Cai, W, Peng, S, Xu, T, Tian, Y, Li, Y, Liu, J. | | Deposit date: | 2021-07-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human MICA mutants in complex with natural killer cell receptor NKG2D
to be published
|
|
6JKP
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense in complex with NAD+ | | Descriptor: | Methanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6JKO
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense | | Descriptor: | Methanol dehydrogenase, ZINC ION | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6IXJ
 
 | | The crystal structure of sulfoacetaldehyde reductase from Klebsiella oxytoca | | Descriptor: | 2-hydroxyethylsulfonic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sulfoacetaldehyde reductase | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural investigation of sulfoacetaldehyde reductase fromKlebsiella oxytoca.
Biochem. J., 476, 2019
|
|
7WNH
 
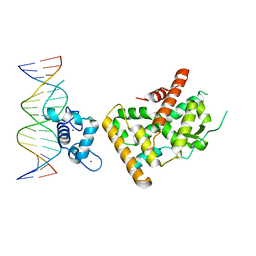 | | Crystal structure of Nurr1 binding to NBRE | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*AP*AP*AP*GP*GP*TP*CP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*GP*AP*CP*CP*TP*TP*TP*TP*CP*GP*G)-3'), Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Zhao, M, Xu, T, Wang, N, Guo, Y, Liu, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Integrative analysis reveals structural basis for transcription activation of Nurr1 and Nurr1-RXR alpha heterodimer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8HQU
 
 | |
5XDZ
 
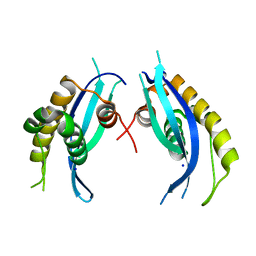 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
5JDR
 
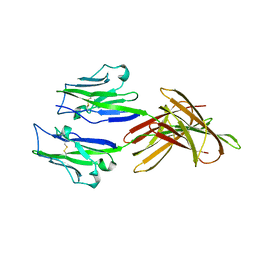 | | Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2016-04-17 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade.
Cell Discov, 3, 2017
|
|
5JDS
 
 | |
5UF7
 
 | | CRYSTAL STRUCTURE OF MUNC13-1 MUN DOMAIN | | Descriptor: | Protein unc-13 homolog A | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2017-01-03 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
7M74
 
 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
6WNH
 
 | | Menin bound to inhibitor M-808 | | Descriptor: | Menin, methyl [(1S,2R)-2-{(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-[1-({1-[4-({1-[4-(piperidin-1-yl)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl]ethyl}cyclopentyl]carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of M-808 as a Highly Potent, Covalent, Small-Molecule Inhibitor of the Menin-MLL Interaction with StrongIn VivoAntitumor Activity.
J.Med.Chem., 63, 2020
|
|
7YK4
 
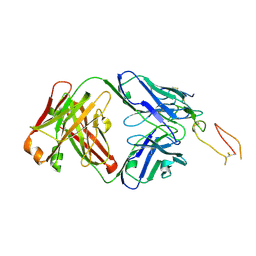 | | ox40-antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 4, antibody-H, ... | | Authors: | Zhou, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
7KW1
 
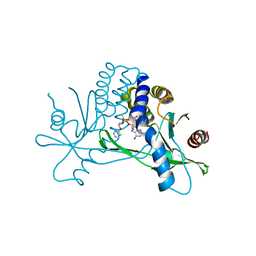 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
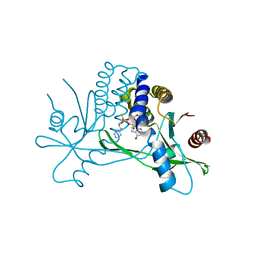 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVZ
 
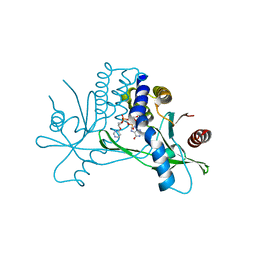 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
