8G9J
 
 | |
8G9K
 
 | |
8GA6
 
 | |
8GA7
 
 | |
8GEL
 
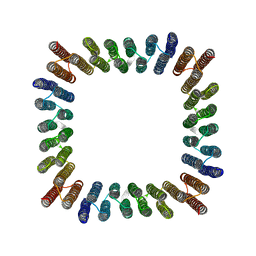 | | Cryo-EM structure of synthetic tetrameric building block sC4 | | Descriptor: | sC4 | | Authors: | Redler, R.L, Huddy, T.F, Hsia, Y, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Blueprinting extendable nanomaterials with standardized protein blocks.
Nature, 627, 2024
|
|
2YMK
 
 | |
7U62
 
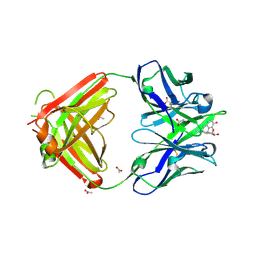 | | Crystal structure of Anti-Heroin Antibody HY4-1F9 Fab Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, ACETATE ION, HY4-1F9 Fab Heavy Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
7U64
 
 | |
7U61
 
 | | Crystal Structure of Anti-Nicotine Antibody NIC311 Fab Complexed with Nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, NIC311 Fab Heavy Chain, NIC311 Fab Light Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P, Liban, T.L. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
6OKO
 
 | |
5MBW
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH Pep#3 | | Descriptor: | BACE1 INHIBITOR PEPTIDE Pep#3, Beta-secretase 1, CHLORIDE ION | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCO
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
7ALO
 
 | | Structure of B*27:09/photoRL9 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loll, B, Lan, H, Freund, C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exchange catalysis by tapasin exploits conserved and allele-specific features of MHC-I molecules.
Nat Commun, 12, 2021
|
|
8FAR
 
 | |
4LSZ
 
 | | Caspase-7 in Complex with DARPin D7.18 | | Descriptor: | Caspase-7 subunit p10, Caspase-7 subunit p20, DARPin D7.18 | | Authors: | Fluetsch, A, Lukarska, M, Gruetter, M.G. | | Deposit date: | 2013-07-23 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combined inhibition of caspase 3 and caspase 7 by two highly selective DARPins slows down cellular demise.
Biochem.J., 461, 2014
|
|
6SHK
 
 | |
6SNJ
 
 | |
8UNG
 
 | |
1TFX
 
 | |
3BJM
 
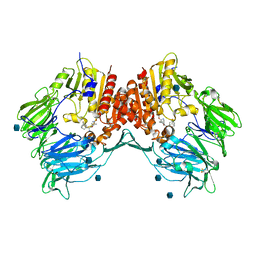 | | Crystal structure of human DPP-IV in complex with (1S,3S, 5S)-2-[(2S)-2-AMINO-2-(3-HYDROXYTRICYCLO[3.3.1.13,7]DEC-1- YL)ACETYL]-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (CAS), (1S,3S,5S)-2-((2S)-2-AMINO-2-(3-HYDROXYADAMANTAN-1- YL)ACETYL)-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (IUPAC), OR BMS-477118 | | Descriptor: | (2~{S})-2-azanyl-1-[(1~{S},3~{S},5~{S})-3-(iminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(5~{R},7~{S})-3-oxidanyl-1-ad amantyl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2007-12-04 | | Release date: | 2008-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Involvement of DPP-IV catalytic residues in enzyme-saxagliptin complex formation.
Protein Sci., 17, 2008
|
|
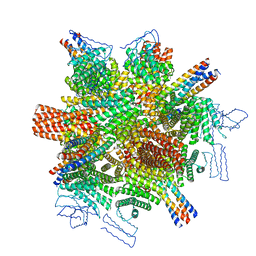
8CWY
 
 | |
8CUS
 
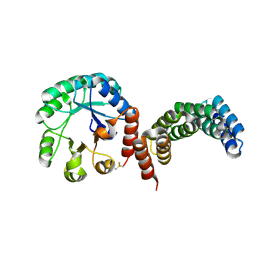 | |
8CUT
 
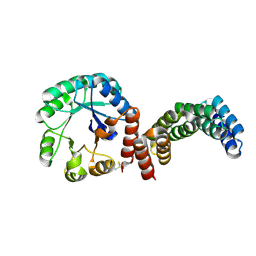 | |
8CUW
 
 | |
