7CT4
 
 | | Crystal structure of D-amino acid oxidase from Rasamsonia emersonii strain YA | | Descriptor: | D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Shimekake, Y, Hirato, Y, Okazaki, S, Funabashi, R, Goto, M, Furuichi, T, Suzuki, H, Takahashi, S. | | Deposit date: | 2020-08-18 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of a unique D-amino-acid oxidase from the thermophilic fungus Rasamsonia emersonii strain YA.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7Y4K
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-ornitine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-5-(phenylmethoxycarbonylamino)pentanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
7Y4M
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-lysine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-6-(phenylmethoxycarbonylamino)hexanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
5YBW
 
 | | Crystal structure of pyridoxal 5'-phosphate-dependent aspartate racemase | | Descriptor: | Aspartate racemase | | Authors: | Mizobuchi, T, Nonaka, R, Yoshimura, M, Abe, K, Takahashi, S, Kera, Y, Goto, M. | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a pyridoxal 5'-phosphate-dependent aspartate racemase derived from the bivalve mollusc Scapharca broughtonii
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
1VEF
 
 | | Acetylornithine aminotransferase from Thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Three-Dimensional Strutcure of Acetylornithine aminotransferase from Thermus thermophilus HB8
To be Published
|
|
1WKH
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, Acetylornithine/acetyl-lysine aminotransferase | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1WKG
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, N~2~-ACETYL-N~5~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-ORNITHINE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1VGN
 
 | | Structure-based design of the irreversible inhibitors to metallo--lactamase (IMP-1) | | Descriptor: | 3-OXO-3-[(3-OXOPROPYL)SULFANYL]PROPANE-1-THIOLATE, ACETIC ACID, Beta-lactamase IMP-1, ... | | Authors: | Kurosaki, H, Yamaguchi, Y, Higashi, T, Soga, K, Matsueda, S, Misumi, S, Yamagata, Y, Arakawa, Y, Goto, M. | | Deposit date: | 2004-04-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Irreversible Inhibition of Metallo-beta-lactamase (IMP-1) by 3-(3-Mercaptopropionylsulfanyl)propionic Acid Pentafluorophenyl Ester
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1WUP
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81E) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
1WUO
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81A) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
2DOU
 
 | | probable N-succinyldiaminopimelate aminotransferase (TTHA0342) from Thermus thermophilus HB8 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, probable N-succinyldiaminopimelate aminotransferase | | Authors: | Omi, R, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-03 | | Release date: | 2006-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | probable N-succinyldiaminopimelate aminotransferase (TTHA0342) from Thermus thermophilus HB8
To be published
|
|
2EJV
 
 | | Crystal structure of threonine 3-dehydrogenase complexed with NAD+ | | Descriptor: | L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Omi, R, Yao, T, Goto, M, Miyahara, I, Hirotsu, K. | | Deposit date: | 2007-03-20 | | Release date: | 2008-03-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of threonine 3-dehydrogenase
To be Published
|
|
2EJW
 
 | | Homoserine Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Homoserine dehydrogenase, MAGNESIUM ION, ... | | Authors: | Omi, R, Goto, M, Miyahara, I, Hirotsu, K. | | Deposit date: | 2007-03-21 | | Release date: | 2008-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Homoserine Dehydrogenase from Thermus thermophilus HB8
To be Published
|
|
2DQ4
 
 | | Crystal structure of threonine 3-dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-threonine 3-dehydrogenase, ZINC ION | | Authors: | Omi, R, Yao, T, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of threonine 3-dehydrogenase
To be Published
|
|
1RKR
 
 | | CRYSTAL STRUCTURE OF AZURIN-I FROM ALCALIGENES XYLOSOXIDANS NCIMB 11015 | | Descriptor: | AZURIN-I, COPPER (II) ION | | Authors: | Li, C, Inoue, T, Gotowda, M, Suzuki, S, Yamaguchi, K, Kataoka, K, Kai, Y. | | Deposit date: | 1997-05-17 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of azurin I from the denitrifying bacterium Alcaligenes xylosoxidans NCIMB 11015 at 2.45 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BYO
 
 | | WILD-TYPE PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1BYP
 
 | | E43K,D44K DOUBLE MUTANT PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1BQ5
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS GIFU 1051 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Inoue, T, Gotowda, M, Deligeer, Suzuki, S, Kataoka, K, Yamaguchi, K, Watanabe, H, Goho, M, Yasushi, K.A.I. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 A resolution: comparison of blue and green nitrite reductases.
J.Biochem.(Tokyo), 124, 1998
|
|
1KDJ
 
 | | OXIDIZED FORM OF PLASTOCYANIN FROM DRYOPTERIS CRASSIRHIZOMA | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Gotowda, M, Hamada, K, Kohzuma, T, Yoshizaki, F, Sugimura, Y, Kai, Y. | | Deposit date: | 1998-05-08 | | Release date: | 1999-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure and unusual pH dependence of plastocyanin from the fern Dryopteris crassirhizoma. The protonation of an active site histidine is hindered by pi-pi interactions.
J.Biol.Chem., 274, 1999
|
|
1KDI
 
 | | REDUCED FORM OF PLASTOCYANIN FROM DRYOPTERIS CRASSIRHIZOMA | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Inoue, T, Gotowda, M, Hamada, K, Kohzuma, T, Yoshizaki, F, Sugimura, Y, Kai, Y. | | Deposit date: | 1998-05-08 | | Release date: | 1999-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and unusual pH dependence of plastocyanin from the fern Dryopteris crassirhizoma. The protonation of an active site histidine is hindered by pi-pi interactions.
J.Biol.Chem., 274, 1999
|
|
1VC3
 
 | |
6AHS
 
 | | Mouse Kallikrein 7 in complex with imidazolinylindole derivative | | Descriptor: | 1-[(2-chlorophenyl)sulfonyl]-5-methyl-3-[(4R)-2-methyl-4,5-dihydro-1H-imidazol-4-yl]-1H-indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and structure-activity relationship of imidazolinylindole derivatives as kallikrein 7 inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
8GZC
 
 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
6ANL
 
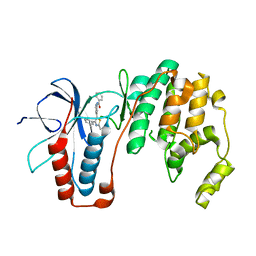 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, TAK-715 | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-08-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
8H4V
 
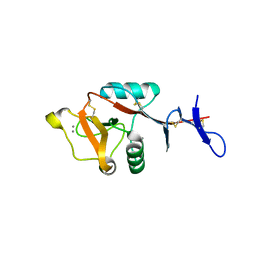 | | Mincle CRD complex with PGL trisaccharide | | Descriptor: | (2~{R},3~{R},4~{S},5~{S},6~{R})-6-(methoxymethyl)oxane-2,3,4,5-tetrol-(1-4)-6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-3-O-methyl-alpha-L-rhamnopyranose, C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Ishizuka, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PGL-III, a Rare Intermediate of Mycobacterium leprae Phenolic Glycolipid Biosynthesis, Is a Potent Mincle Ligand.
Acs Cent.Sci., 9, 2023
|
|
