8F5P
 
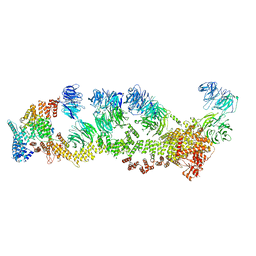 | | Structure of Leishmania tarentolae IFT-A (state 2) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
8F5O
 
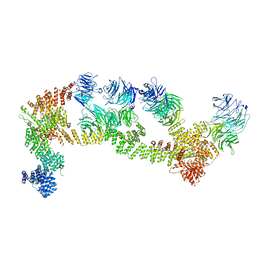 | | Structure of Leishmania tarentolae IFT-A (state 1) | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 122B, putative, ... | | Authors: | Zhou, H, Brown, A. | | Deposit date: | 2022-11-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of IFT-A polymerization into trains for ciliary transport.
Cell, 185, 2022
|
|
3VZD
 
 | | Crystal structure of Sphingosine Kinase 1 with inhibitor and ADP | | Descriptor: | 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
3VZB
 
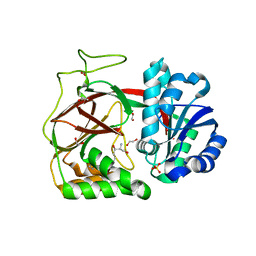 | | Crystal structure of Sphingosine Kinase 1 | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
3VZC
 
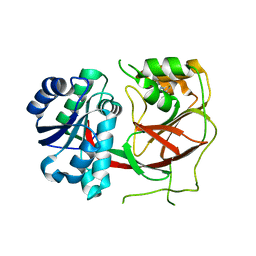 | | Crystal structure of Sphingosine Kinase 1 with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-(4-chlorophenyl)-1,3-thiazol-2-yl]amino}phenol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-10-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of sphingosine kinase 1 substrate recognition and catalysis.
Structure, 21, 2013
|
|
1RDF
 
 | | G50P mutant of phosphonoacetaldehyde hydrolase in complex with substrate analogue vinyl sulfonate | | Descriptor: | ETHANESULFONIC ACID, MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-11-05 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of the substrate specificity loop of the HAD superfamily cap domain
Biochemistry, 43, 2004
|
|
5JRS
 
 | |
2FUC
 
 | | Human alpha-Phosphomannomutase 1 with Mg2+ cofactor bound | | Descriptor: | MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
2FUE
 
 | | Human alpha-Phosphomannomutase 1 with D-mannose 1-phosphate and Mg2+ cofactor bound | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, Phosphomannomutase 1 | | Authors: | Silvaggi, N.R, Zhang, C, Lu, Z, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2006-01-26 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a.
J.Biol.Chem., 281, 2006
|
|
6O2L
 
 | |
2LBY
 
 | |
6J1U
 
 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
1ZVA
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZVB
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | E2 glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZV8
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | ACETATE ION, CACODYLATE ION, E2 glycoprotein, ... | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
1ZV7
 
 | | A structure-based mechanism of SARS virus membrane fusion | | Descriptor: | CHLORIDE ION, spike glycoprotein | | Authors: | Deng, Y, Liu, J, Zheng, Q, Yong, W, Dai, J, Lu, M. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the SARS Virus S2 Protein.
Structure, 14, 2006
|
|
6JJ0
 
 | |
6JQH
 
 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | Authors: | Liu, Z, Yan, A, Gao, Y. | | Deposit date: | 2022-10-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
1RYG
 
 | |
1RYV
 
 | |
8ITE
 
 | |
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7E35
 
 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
