1V1H
 
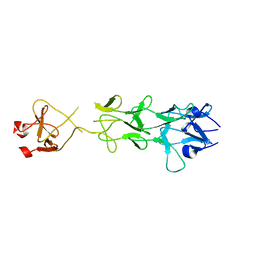 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a short linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
1V1I
 
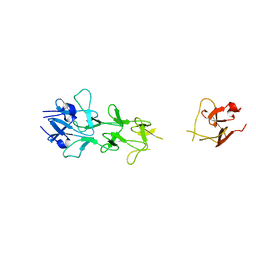 | | Adenovirus fibre shaft sequence N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif with a long linker | | Descriptor: | FIBRITIN, FIBER PROTEIN | | Authors: | Papanikolopoulou, K, Teixeira, S, Belrhali, H, Forsyth, V.T, Mitraki, A, van Raaij, M.J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-07-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenovirus Fibre Shaft Sequences Fold Into the Native Triple Beta-Spiral Fold When N-Terminally Fused to the Bacteriophage T4 Fibritin Foldon Trimerisation Motif
J.Mol.Biol., 342, 2004
|
|
8RCU
 
 | | human TRPM4 in SMA IBA | | Descriptor: | 4-chloranyl-2-[2-(3-iodanylphenoxy)ethanoylamino]benzoic acid, CHOLESTEROL, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Ekundayo, B, Prakash, A, Christian, G, Anne, F.H, Sabrina, G, Mey, B, Daniela, R, Martin, L, Jean, S.R, Henning, S, Hugues, A, Dongchun, N. | | Deposit date: | 2023-12-07 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a Binding Site for Small Molecule Inhibitors Targeting Human
To Be Published
|
|
8RD9
 
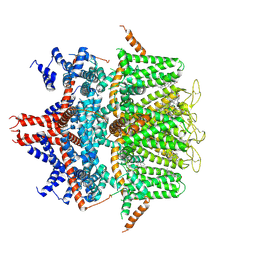 | | human TRPM4 in SMA NBA | | Descriptor: | 4-chloranyl-2-(2-naphthalen-1-yloxyethanoylamino)benzoic acid, CHOLESTEROL, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Ekundayo, B, Prakash, A, Christian, G, Anne, F.H, Sabrina, G, Mey, B, Daniela, R, Martin, L, Jean, S.R, Henning, S, Hugues, A, Dongchun, N. | | Deposit date: | 2023-12-07 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Identification of a Binding Site for Small Molecule Inhibitors Targeting Human
To Be Published
|
|
8RCR
 
 | | human TRPM4 in SMA apo | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Ekundayo, B, Prakash, A, Christian, G, Anne, F.H, Sabrina, G, Mey, B, Daniela, R, Martin, L, Jean, S.R, Henning, S, Hugues, A, Dongchun, N. | | Deposit date: | 2023-12-07 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Identification of a Binding Site for Small Molecule Inhibitors Targeting Human
To Be Published
|
|
9EPR
 
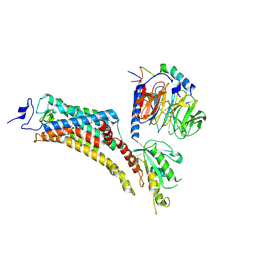 | | Cryo-EM Structure of Jumping Spider Rhodopsin-1 bound to a Gi heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tejero, O, Pamula, F, Koyanagi, M, Nagata, T, Afanasyev, P, Das, I, Deupi, X, Sheves, M, Terakita, A, Schertler, G.F.X, Rodrigues, M.J, Tsai, C.-J. | | Deposit date: | 2024-03-19 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Active state structures of a bistable visual opsin bound to G proteins.
Nat Commun, 15, 2024
|
|
3JU0
 
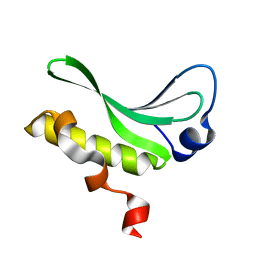 | | Structure of the arm-type binding domain of HAI7 integrase | | Descriptor: | Phage integrase | | Authors: | Szwagierczak, A, Antonenka, U, Popowicz, G.M, Sitar, T, Holak, T.A, Rakin, A. | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the arm-type binding domains of HPI and HAI7 integrases
J.Biol.Chem., 284, 2009
|
|
7A5U
 
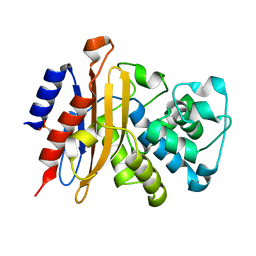 | |
7A74
 
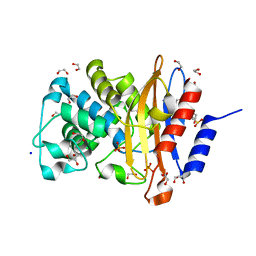 | | Structure of G132N BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two beta-Lactamase Variants with Reduced Clavulanic Acid Inhibition Display Different Millisecond Dynamics.
Antimicrob.Agents Chemother., 65, 2021
|
|
5MRB
 
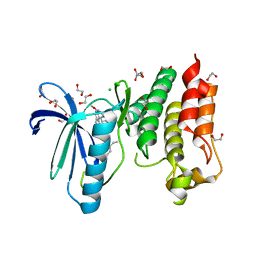 | | Crystal structure of human Mps1 (TTK) in complex with Cpd-5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dual specificity protein kinase TTK, ... | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-12-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
7A72
 
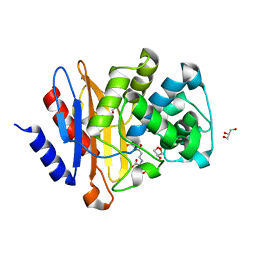 | | Structure of G132S BlaC from Mycobacterium tuberculosis bound to the trans-enamine adduct of sulbactam | | Descriptor: | ACETATE ION, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
7A5W
 
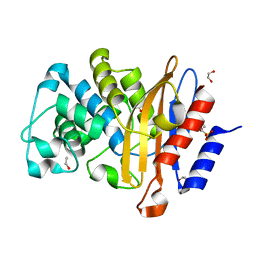 | | Structure of D172N BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-24 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
7A71
 
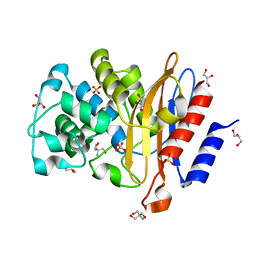 | | Structure of G132S BlaC from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
7A5T
 
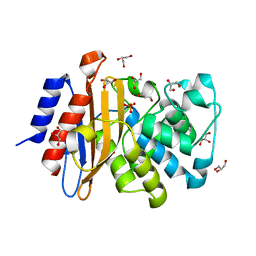 | | Crystal structure of A55E mutant of BlaC from Mycobacterium tuberculosis | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-21 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
7A6Z
 
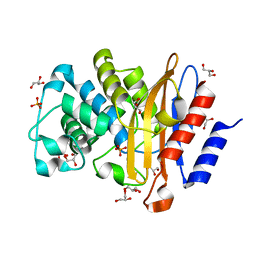 | | Structure of P226G BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved proline residues prevent dimerization and aggregation in the beta-lactamase BlaC.
Protein Sci., 33, 2024
|
|
1OH5
 
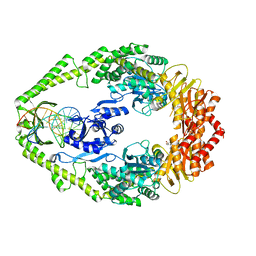 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A C:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*CP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH6
 
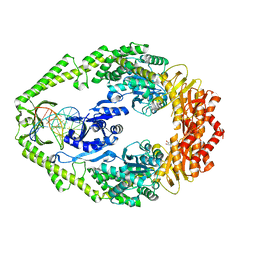 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN A:A MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*AP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP* CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*AP*TP*GP*GP* CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH8
 
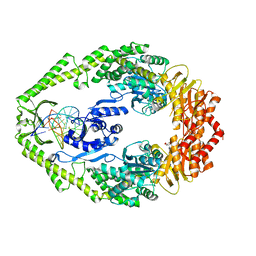 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH AN UNPAIRED THYMIDINE | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP* TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*CP*TP*TP*GP*GP*CP* AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
1OH7
 
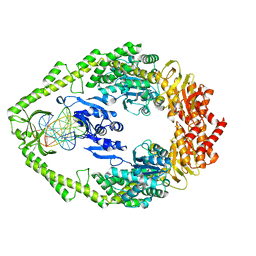 | | THE CRYSTAL STRUCTURE OF E. COLI MUTS BINDING TO DNA WITH A G:G MISMATCH | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP *CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP *GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*GP*TP*GP*GP *CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Lamers, M.H, Enzlin, J.H, Winterwerp, H.H.K, Perrakis, A, Sixma, T.K. | | Deposit date: | 2003-05-23 | | Release date: | 2003-08-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of E. Coli DNA Mismatch Repair Enzyme Muts in Complex with Different Mismatches: A Common Recognition Mode for Diverse Substrates
Nucleic Acids Res., 31, 2003
|
|
7P0K
 
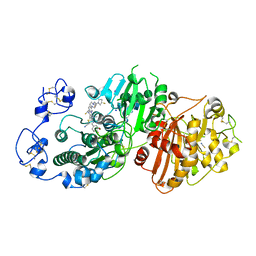 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
5O91
 
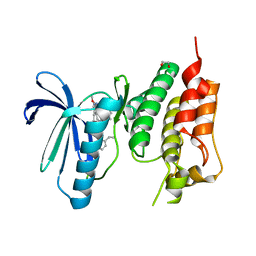 | | Crystal structure of human Mps1 (TTK) C604W mutant in complex with Cpd-5 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, ~{N}-(2,6-diethylphenyl)-8-[[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino]-1-methyl-4,5-dihydropyrazolo[4,3-h]quinazoline-3-carboxamide | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2017-06-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
5NTT
 
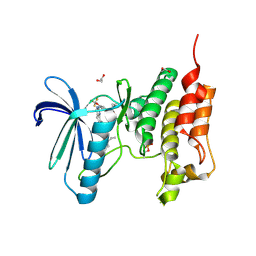 | | Crystal structure of human Mps1 (TTK) C604Y mutant in complex with NMS-P715 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N-(2,6-DIETHYLPHENYL)-1-METHYL-8-({4-[(1-METHYLPIPERIDIN-4-YL)CARBAMOYL]-2-(TRIFLUOROMETHOXY)PHENYL}AMINO)-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
5LJJ
 
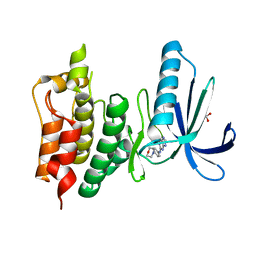 | | Crystal structure of human Mps1 (TTK) in complex with Reversine | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2016-07-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of reversine selectivity in inhibiting Mps1 more potently than aurora B kinase.
Proteins, 84, 2016
|
|
5M0D
 
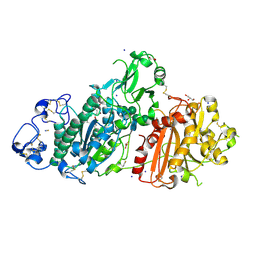 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2,Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, GLYCEROL, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
5M0E
 
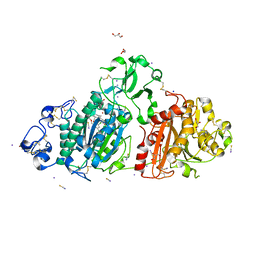 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | 7alpha-hydroxycholesterol, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
