4AH6
 
 | | Human mitochondrial aspartyl-tRNA synthetase | | Descriptor: | ASPARTATE--TRNA LIGASE, MITOCHONDRIAL | | Authors: | Neuenfeldt, A, Sissler, M, Lorber, B, Florentz, C, Sauter, C. | | Deposit date: | 2012-02-03 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Thermodynamic Properties Distinguish Human Mitochondrial Aspartyl-tRNA Synthetase from Bacterial Homolog with Same 3D Architecture
Nucleic Acids Res., 41, 2013
|
|
7NZ7
 
 | | Crystal structure of mouse ADAT2/ADAT3 tRNA deamination complex 1 | | Descriptor: | Probable inactive tRNA-specific adenosine deaminase-like protein 3, ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Ramos Morales, E, Romier, C. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structure of the mouse ADAT2/ADAT3 complex reveals the molecular basis for mammalian tRNA wobble adenosine-to-inosine deamination.
Nucleic Acids Res., 49, 2021
|
|
7NZ9
 
 | |
7NZ8
 
 | | Crystal structure of mouse ADAT2/ADAT3 tRNA deamination complex 2 | | Descriptor: | Probable inactive tRNA-specific adenosine deaminase-like protein 3, ZINC ION, tRNA-specific adenosine deaminase 2 | | Authors: | Ramos Morales, E, Romier, C. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The structure of the mouse ADAT2/ADAT3 complex reveals the molecular basis for mammalian tRNA wobble adenosine-to-inosine deamination.
Nucleic Acids Res., 49, 2021
|
|
5D99
 
 | | 3DW4 redetermined by direct methods starting from random phase angles | | Descriptor: | GLYCEROL, RNA (27-MER) hairpin from sarcin-ricin domain of E. coli 23S rRNA | | Authors: | Mooers, B.H.M. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Direct-methods structure determination of a trypanosome RNA-editing substrate fragment with translational pseudosymmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6HSG
 
 | | Crystal structure of Schistosoma mansoni HDAC8 H292M mutant complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
8CEF
 
 | | Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA | | Descriptor: | DNA (26-MER), Nuclear receptor DNA binding domain, ZINC ION | | Authors: | Patel, A.K.M, Shaik, T.B, McEwen, A.G, Moras, D, Klaholz, B.P, Billas, I.M.L. | | Deposit date: | 2023-02-01 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Nucleic Acids Res., 51, 2023
|
|
8P0A
 
 | | Human Cohesin ATPase module | | Descriptor: | 64-kDa C-terminal product, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Landwerlin, P, Durand, A, Diebold-Durand, M.-L, Romier, C. | | Deposit date: | 2023-05-10 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
3ITH
 
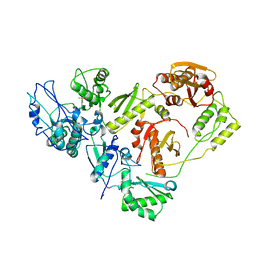 | | Crystal structure of the HIV-1 reverse transcriptase bound to a 6-vinylpyrimidine inhibitor | | Descriptor: | 6-ethenyl-N,N-dimethyl-2-(methylsulfonyl)pyrimidin-4-amine, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Freisz, S, Bec, G, Wolff, P, Dumas, P, Radi, M, Botta, M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of HIV-1 Reverse Transcriptase Bound to a Non-Nucleoside Inhibitor with a Novel Mechanism of Action
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
8PQ5
 
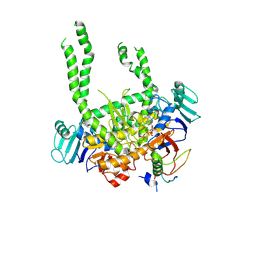 | | Human Cohesin ATPase module with an open DNA exit gate | | Descriptor: | 64-kDa C-terminal product, ADENOSINE-5'-TRIPHOSPHATE, Structural maintenance of chromosomes protein 1A, ... | | Authors: | Landwerlin, P, Durand, A, Diebold-Durand, M.-L, Romier, C. | | Deposit date: | 2023-07-10 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
5OB3
 
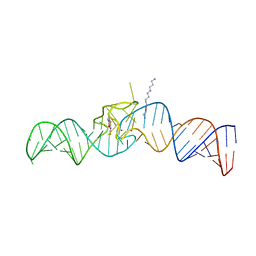 | | iSpinach aptamer | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, POTASSIUM ION, RNA aptamer (69-MER), ... | | Authors: | Fernandez-Millan, P, Autour, A, Westhof, E, Ryckelynck, M. | | Deposit date: | 2017-06-26 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure and fluorescence properties of the iSpinach aptamer in complex with DFHBI.
RNA, 23, 2017
|
|
8ROB
 
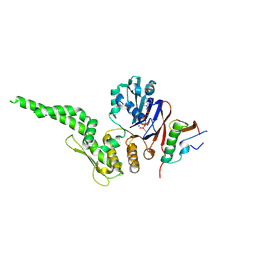 | |
8ROF
 
 | |
8RO7
 
 | |
8RO8
 
 | |
8RO9
 
 | | Human cohesin SMC1A-HD(longCC-EQ)/RAD21-C complex - Open/closed P-loop conformation | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 64-kDa C-terminal product, Structural maintenance of chromosomes protein 1A | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROC
 
 | |
8ROE
 
 | |
8ROG
 
 | |
8ROJ
 
 | | Human cohesin SMC3-HD/RAD21-N complex - ADP-Mg-bound conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROI
 
 | | Human cohesin SMC3-HD(EQ)/RAD21-N complex - ADP-bound conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Double-strand-break repair protein rad21 homolog, Structural maintenance of chromosomes protein 3 | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROA
 
 | |
8ROH
 
 | | Human cohesin SMC3-HD(EQ)/RAD21-N complex - Apo conformation | | Descriptor: | Double-strand-break repair protein rad21 homolog, GLYCEROL, SULFATE ION, ... | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROL
 
 | |
8RO6
 
 | |
