6D00
 
 | | Calcarisporiella thermophila Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcarisporiella thermophila Hsp104 | | Authors: | Zhang, K, Pintilie, G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
7P2B
 
 | |
6VLK
 
 | | A varicella-zoster virus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, ... | | Authors: | Xing, Y. | | Deposit date: | 2020-01-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4529 Å) | | Cite: | A glycoprotein B-neutralizing antibody structure at 2.8 angstrom uncovers a critical domain for herpesvirus fusion initiation.
Nat Commun, 11, 2020
|
|
6VN1
 
 | |
3QM0
 
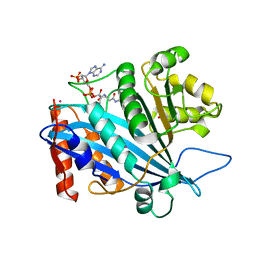 | | Crystal structure of RTT109-AC-CoA complex | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | Authors: | Tang, Y, Marmorstein, R. | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5DBM
 
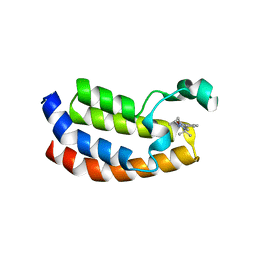 | | Crystal structure of the CBP bromodomain in complex with CPI703 | | Descriptor: | (4R)-6-(1-tert-butyl-1H-pyrazol-4-yl)-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, CREB-binding protein | | Authors: | Setser, J.W, Poy, F, Bellon, S.F. | | Deposit date: | 2015-08-21 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Regulatory T Cell Modulation by CBP/EP300 Bromodomain Inhibition.
J.Biol.Chem., 291, 2016
|
|
6BAO
 
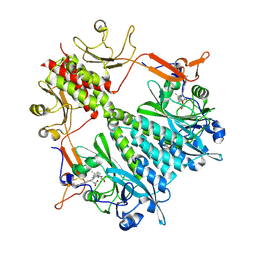 | | Stigmatella aurantiaca phytochrome photosensory core module, wild type | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
6BAK
 
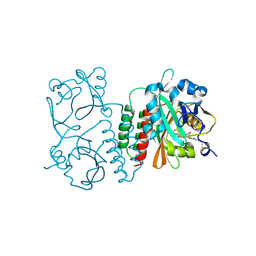 | | The structure of the Stigmatella aurantiaca phytochrome chromophore binding domain T289H mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Schmidt, M, Stojkovic, E. | | Deposit date: | 2017-10-13 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for light control of cell development revealed by crystal structures of a myxobacterial phytochrome.
IUCrJ, 5, 2018
|
|
4GFX
 
 | | Crystal structure of the N-terminal domain of TXNIP | | Descriptor: | GLYCEROL, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2012-08-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein.
Nat Commun, 5, 2014
|
|
4HBM
 
 | | Ordering of the N Terminus of Human MDM2 by Small Molecule Inhibitors | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S)-1-hydroxybutan-2-yl]-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ordering of the N-terminus of human MDM2 by small molecule inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
8GJI
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | GCG binder, Glucagon | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GJG
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
7OPM
 
 | | Phosphorylated ERK2 in complex with ORF45 | | Descriptor: | 1-[4-(hydroxymethyl)-1H-pyrazolo[4,3-c]pyridin-6-yl]-3-[(1S)-1-phenylethyl]urea, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sok, P, Remenyi, A, Alexa, A, Poti, A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host.
Nat Commun, 13, 2022
|
|
7OPO
 
 | | RSK2 N-terminal kinase domain in complex with ORF45 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein ORF45, Ribosomal protein S6 kinase alpha-3 | | Authors: | Sok, P, Remenyi, A, Alexa, A, Poti, A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host.
Nat Commun, 13, 2022
|
|
7U9S
 
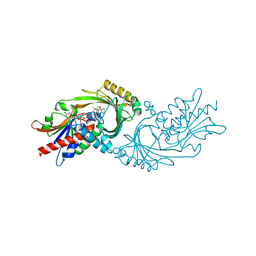 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | 5-{2-[4-(trifluoromethyl)phenyl]ethyl}-1,4-dihydropyrazine-2,3-dione, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7U9U
 
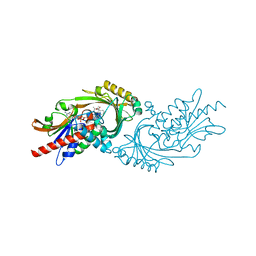 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | (3R)-3-(5,6-dioxo-1,4,5,6-tetrahydropyrazin-2-yl)-2,3-dihydro-1,4-benzoxathiine-7-carbonitrile, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7TTR
 
 | | Skd3_ATPyS_FITC-casein Hexamer, AAA+ only | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-casein, Caseinolytic peptidase B protein homolog, ... | | Authors: | Rizo, A.N. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unique structural features govern the activity of a human mitochondrial AAA+ disaggregase, Skd3.
Cell Rep, 40, 2022
|
|
7TTS
 
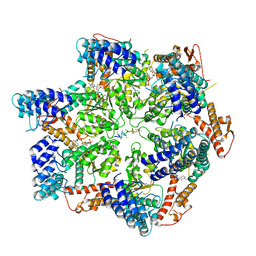 | | Skd3, hexamer, filtered | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-casein, Caseinolytic peptidase B protein homolog, ... | | Authors: | Rizo, A.N, Cupo, R.R. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Unique structural features govern the activity of a human mitochondrial AAA+ disaggregase, Skd3.
Cell Rep, 40, 2022
|
|
5YI4
 
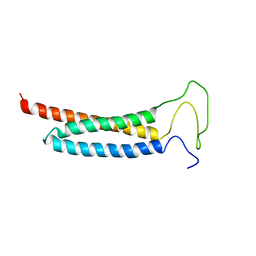 | | Solution Structure of the DISC1/Ndel1 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Nuclear distribution protein nudE-like 1 | | Authors: | Ye, F, Yu, C, Yu, C, Zhang, M. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | DISC1 Regulates Neurogenesis via Modulating Kinetochore Attachment of Ndel1/Nde1 during Mitosis.
Neuron, 96, 2017
|
|
8F4S
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
6FS1
 
 | | MCL1 in complex with an indole acid ligand | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Kasmirski, S, Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS2
 
 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
6FS0
 
 | |
8GSA
 
 | |
