6RT6
 
 | |
5ZFZ
 
 | | Crystal structure of human DUX4 homeodomains bound to A12T DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*TP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*AP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5ZFW
 
 | | Crystal structure of human DUX4 homeodomains bound to A11G DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*GP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*CP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5ZFY
 
 | | Crystal structure of human DUX4 homeodomains bound to A12C DNA mutant | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*CP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*GP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4-like protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
5Z6Z
 
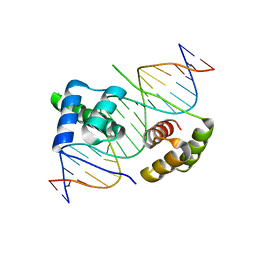 | | Crystal structure of human DUX4 homeodomains bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*CP*TP*AP*AP*CP*CP*TP*AP*AP*TP*CP*AP*CP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*GP*TP*GP*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*TP*GP*G)-3'), Double homeobox protein 4 | | Authors: | Li, Y.Y, Wu, B.X, Gan, J.H. | | Deposit date: | 2018-01-26 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for multiple gene regulation by human DUX4.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
6YKJ
 
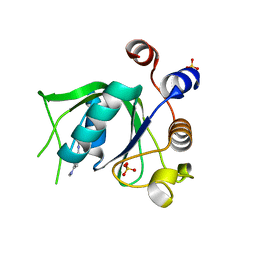 | | Crystal structure of YTHDC1 with compound DHU_DC1_125 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-4-(methylamino)pyridine-2-carboxamide | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNP
 
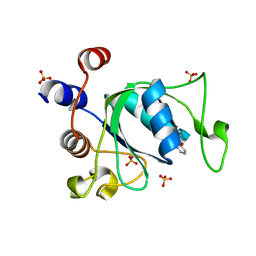 | | Crystal structure of YTHDC1 with compound T96C1 | | Descriptor: | CHLORIDE ION, SULFATE ION, YTHDC1, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNI
 
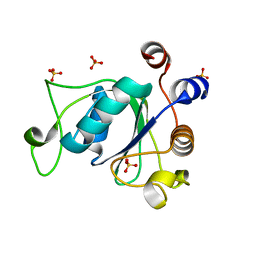 | | Crystal structure of YTHDC1 with compound DHU_DC1_036 | | Descriptor: | (2~{R},3~{R})-2-(3-chlorophenyl)-3,5,5-trimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNL
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_078 | | Descriptor: | 3-[(2~{R})-5,5-dimethyl-2-phenyl-morpholin-4-yl]-~{N}-(methylcarbamoyl)propanamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YL8
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_011 | | Descriptor: | (2~{R},5~{S})-2-(2-chlorophenyl)-5-methyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YNO
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_139 | | Descriptor: | 6-[[methyl-[(1-phenylpyrazol-3-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD9
 
 | | Crystal structure of YTHDC1 apo purified using GST tag | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Caflisch, A. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZCN
 
 | | Crystal structure of YTHDC1 with m6A | | Descriptor: | N6-METHYLADENOSINE-5'-MONOPHOSPHATE, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZOT
 
 | |
6ZCM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_180 | | Descriptor: | 6-[[cyclopropyl-[(7-methoxy-1,3-benzodioxol-5-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
1TMJ
 
 | | Crystal structure of E.coli apo-HPPK(W89A) at 1.45 Angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2004-06-10 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Is the Critical Role of Loop 3 of Escherichia coli 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase in Catalysis Due to Loop-3 Residues Arginine-84 and Tryptophan-89? Site-Directed Mutagenesis, Biochemical, and Crystallographic Studies.
Biochemistry, 44, 2005
|
|
6K9V
 
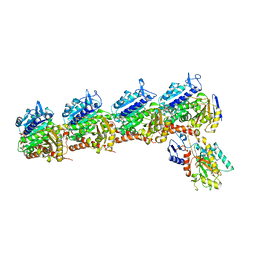 | | Crystal structure of tubulin in complex with inhibitor D64 | | Descriptor: | (5-methoxy-1H-indol-2-yl)-phenyl-methanone, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Structural insights into the design of indole derivatives as tubulin polymerization inhibitors.
Febs Lett., 594, 2020
|
|
6KIH
 
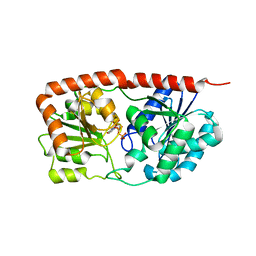 | | Sucrose-phosphate synthase (tll1590) from Thermosynechococcus elongatus | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Tll1590 protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J. | | Deposit date: | 2019-07-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Co-crystal Structure ofThermosynechococcus elongatusSucrose Phosphate Synthase With UDP and Sucrose-6-Phosphate Provides Insight Into Its Mechanism of Action Involving an Oxocarbenium Ion and the Glycosidic Bond.
Front Microbiol, 11, 2020
|
|
8I92
 
 | | ACE2-B0AT1 complex bound with glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
6LDQ
 
 | |
8I91
 
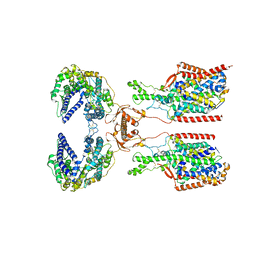 | | ACE2-SIT1 complex bound with proline | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8I93
 
 | | ACE2-B0AT1 complex bound with methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Zhang, Y.Y, Shen, Y.P, Yan, R.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B 0 AT1 and ACE2-SIT1.
Cell Discov, 9, 2023
|
|
8SUX
 
 | | Structure of E. coli PtuA hexamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EEA
 
 | | Structure of E.coli Septu (PtuAB) complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EE7
 
 | |
