3B20
 
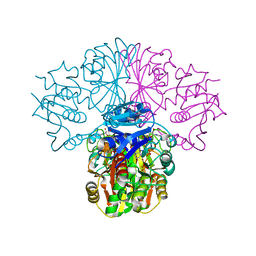 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with NADfrom Synechococcus elongatus" | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Matsumura, H, Kai, A, Maeda, T, Inoue, T. | | Deposit date: | 2011-07-17 | | Release date: | 2012-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
6HZN
 
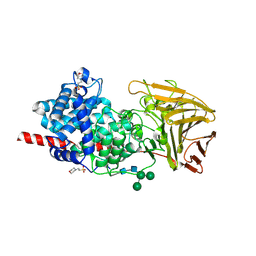 | | Crystal structure of human dermatan sulfate epimerase 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hasan, M, Unge, J, Westergren-Thorsson, G, Ellervik, U, Mueller, U, Malmstrom, A, Tykesson, E. | | Deposit date: | 2018-10-23 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of human dermatan sulfate epimerase 1 emphasizes the importance of C5-epimerization of glucuronic acid in higher organisms
Chem Sci, 2020
|
|
5MGC
 
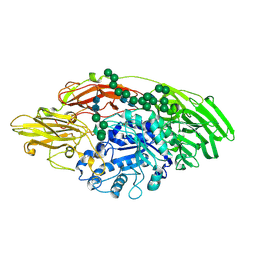 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 4-Galactosyl-lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Probable beta-galactosidase A, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
6I0I
 
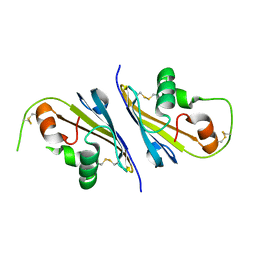 | |
6T5C
 
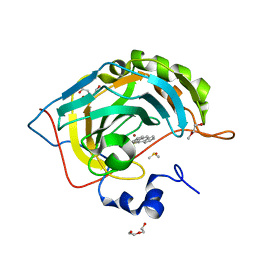 | | Human Carbonic anhydrase II bound by anthracene-9-sulfonamide | | Descriptor: | BICINE, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-10-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Isoform-Selective Enzyme Inhibitors by Exploring Pocket Size According to the Lock-and-Key Principle.
Biophys.J., 119, 2020
|
|
2WAO
 
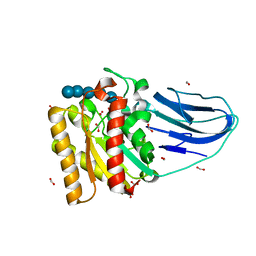 | | Structure of a family two carbohydrate esterase from Clostridium thermocellum in complex with cellohexaose | | Descriptor: | ENDOGLUCANASE E, FORMIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-10 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
6T97
 
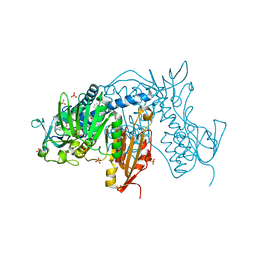 | | Trypanothione Reductase from Leismania infantum in complex with TRL190 | | Descriptor: | 4-[1-[4-[4-(2-phenylethyl)-1,3-thiazol-2-yl]-3-(2-piperidin-4-ylethoxy)phenyl]-1,2,3-triazol-4-yl]butan-1-amine, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carriles, A, Hermoso, J.A. | | Deposit date: | 2019-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffold hopping identifies new triazole-and triazolium-based inhibitors of Leishmania infantum Trypanothione Reductase with potent and selective antileishmanial activity
To Be Published
|
|
4J1E
 
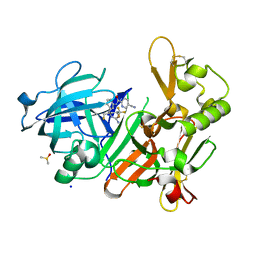 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Cyano-pyridine-2-carboxylic acid [3-((4S,6S)-2-amino-4-fluoromethyl-6-trifluoromethyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, N-{3-[(4S,6S)-2-amino-4-(fluoromethyl)-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-oxazin-4-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
5BKM
 
 | | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-20 | | Release date: | 2022-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal Structure of Hip1 (Rv2224c) mutant - S228DHA (dehydroalanine)
To Be Published
|
|
4ISW
 
 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-11 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
5AP7
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, MONOPOLAR SPINDLE KINASE 1, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.M, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
6YVP
 
 | | Human histidine triad nucleotide-binding protein 2 (hHINT2) complexed with dGMP and refined to 2.77 A | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, Histidine triad nucleotide-binding protein 2, mitochondrial | | Authors: | Dolot, R.D, Krakowiak, A, Nawrot, B.C. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Biochemical, crystallographic and biophysical characterization of histidine triad nucleotide-binding protein 2 with different ligands including a non-hydrolyzable analog of Ap4A.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
6ZJU
 
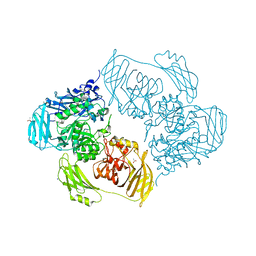 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with saccharose | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
5S4J
 
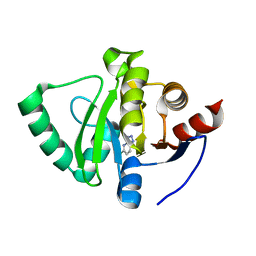 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | Descriptor: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.124 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6V28
 
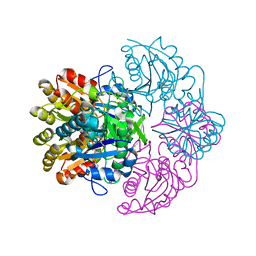 | |
5AP4
 
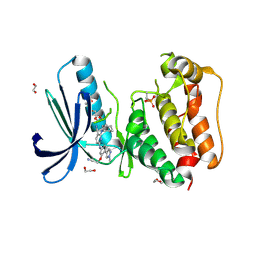 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | Descriptor: | GLYCEROL, Nanobody | | Authors: | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6VFL
 
 | |
5AR5
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Benzimidazole | | Descriptor: | 2-(2-(4-CHLOROPHENYL)-1H-IMIDAZOL-5-YL)-N,1-BIS(2-METHOXYETHYL)-1H-BENZO[D]IMIDAZOLE-5-CARBOXAMIDE, CALCIUM ION, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
4J0T
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-Ethoxy-pyridine-2-carboxylic acid [3-((R)-2-amino-5,5-difluoro-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide | | Descriptor: | 5-Ethoxy-pyridine-2-carboxylic acid [3-((R)-2-amino-5,5-difluoro-4-methyl-5,6-dihydro-4H-[1,3]oxazin-4-yl)-4-fluoro-phenyl]-amide, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | beta-Secretase (BACE1) Inhibitors with High In Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer s Disease
J.Med.Chem., 56, 2013
|
|
5ARD
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, GLYCEROL, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5AR4
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with SB-203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
5AP2
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, ISOPROPYL 6-((4-(1,2-DIMETHYL-1H-IMIDAZOL-5-YL)PHENYL)AMINO)-2-(1-METHYL-1H-PYRAZOL-4-YL)-1H-PYRROLO[3,2-C]PYRIDINE-1-CARBOXYLATE | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
6Z2Q
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (GalGalNAc-TS) product | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Glycodrosocin, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
1YLS
 
 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
