1QQ0
 
 | | COBALT SUBSTITUTED CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-10 | | Release date: | 1999-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
3PV8
 
 | |
3VB1
 
 | |
3V2L
 
 | |
2PFK
 
 | |
3Q79
 
 | |
3Q75
 
 | |
4MR4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MR6
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 2, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MR3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-OH) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1JCS
 
 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE PEPTIDE SUBSTRATE TKCVFM AND AN ANALOG OF FARNESYL DIPHOSPHATE | | Descriptor: | ACETIC ACID, PROTEIN FARNESYLTRANSFERASE, ALPHA SUBUNIT, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3E2X
 
 | | H. influenzae beta-carbonic anhydrase, variant V47A | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Rowlett, R.S, Lee, J. | | Deposit date: | 2008-08-06 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evidence for a bicarbonate "escort" site in Haemophilus influenzae beta-carbonic anhydrase .
Biochemistry, 49, 2010
|
|
3E31
 
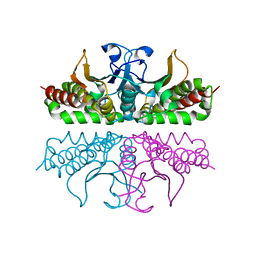 | |
1QRG
 
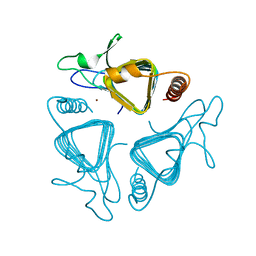 | | A CLOSER LOOK AND THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1JCR
 
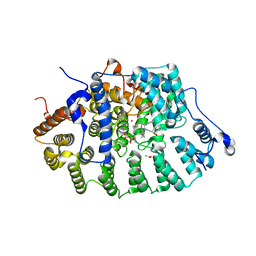 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE NON-SUBSTRATE TETRAPEPTIDE INHIBITOR CVFM AND FARNESYL DIPHOSPHATE SUBSTRATE | | Descriptor: | ACETIC ACID, FARNESYL DIPHOSPHATE, PROTEIN FARNESYLTRANSFERASE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3PX4
 
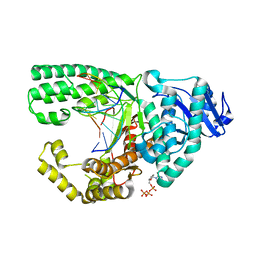 | |
1JCQ
 
 | | CRYSTAL STRUCTURE OF HUMAN PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH FARNESYL DIPHOSPHATE AND THE PEPTIDOMIMETIC INHIBITOR L-739,750 | | Descriptor: | 2(S)-{2(S)-[2(R)-AMINO-3-MERCAPTO]PROPYLAMINO-3(S)-METHYL}PENTYLOXY-3-PHENYLPROPIONYLMETHIONINE SULFONE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4LJN
 
 | | Crystal Structure of MOZ double PHD finger | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
1ZYI
 
 | | Solution structure of ICLN, a multifunctional protein involved in regulatory mechanisms as different as cell volume regulation and rna splicing | | Descriptor: | Methylosome subunit pICln | | Authors: | Fuerst, J, Schedlbauer, A, Gandini, R, Garavaglia, M.L, Siano, S, Gschwentner, M, Sarg, B, Kontaxis, G, Konrat, R, Paulmichl, M. | | Deposit date: | 2005-06-10 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ICln159 folds into a pleckstrin homology domain-like structure. Interaction with kinases and the splicing factor LSm4
J.Biol.Chem., 280, 2005
|
|
4PFK
 
 | | PHOSPHOFRUCTOKINASE. STRUCTURE AND CONTROL | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Evans, P.R, Hudson, P.J. | | Deposit date: | 1988-01-25 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphofructokinase: structure and control.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
3PFK
 
 | |
2ZF9
 
 | |
1JIC
 
 | |
2KHB
 
 | |
