3ICL
 
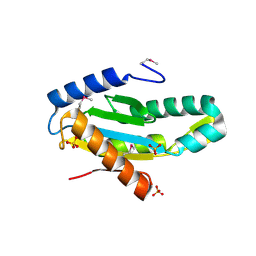 | | X-Ray Structure of Protein (EAL/GGDEF domain protein) from M.capsulatus, Northeast Structural Genomics Consortium Target McR174C | | Descriptor: | EAL/GGDEF domain protein, SULFATE ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target McR174C
To be Published
|
|
3JSR
 
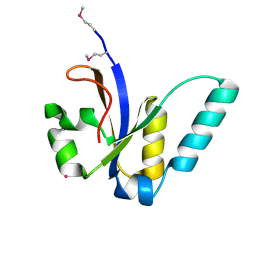 | | X-Ray structure of All0216 protein from Nostoc sp. PCC 7120 at the resolution 1.8A. Northeast Structural Genomics Consortium target NsR236 | | Descriptor: | All0216 protein, POTASSIUM ION | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray structure of All0216 protein from Nostoc sp. PCC 7120 at the resolution 1.8A
To be Published
|
|
4G72
 
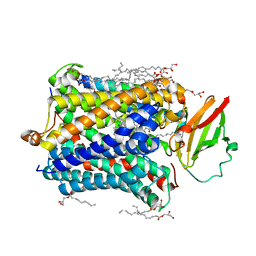 | |
4GP8
 
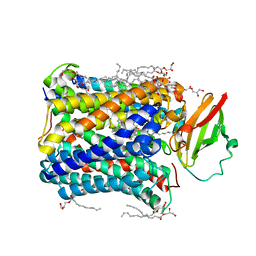 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W+T231F from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
4GP5
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
6M22
 
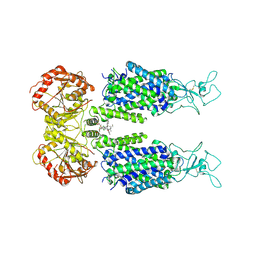 | | KCC3 bound with DIOA | | Descriptor: | 2-[[(2~{R})-2-butyl-6,7-bis(chloranyl)-2-cyclopentyl-1-oxidanylidene-3~{H}-inden-5-yl]oxy]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
6M1Y
 
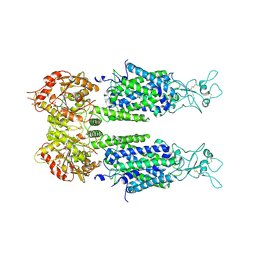 | | The overall structure of KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
7XJW
 
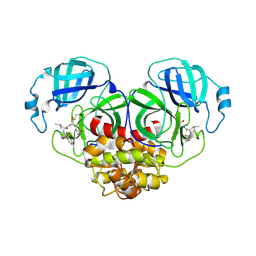 | | Crystal structure of canine coronavirus main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, ORF1a polyprotein | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chiu, Y.F, Chen, Y. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Structural Comparison of SARS-CoV-2 Main Protease and Animal Coronaviral Main Protease Reveals Species-Specific Ligand Binding and Dimerization Mechanism.
Int J Mol Sci, 23, 2022
|
|
2P0Y
 
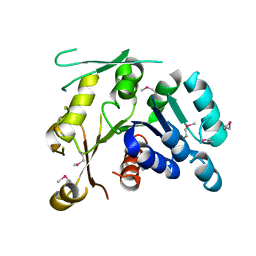 | | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR6 | | Descriptor: | Hypothetical protein lp_0780 | | Authors: | Benach, J, Chen, Y, Seetharaman, J, Chi, K.H, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Q88YI3_LACPL from Lactobacillus plantarum.
To be Published
|
|
8XB1
 
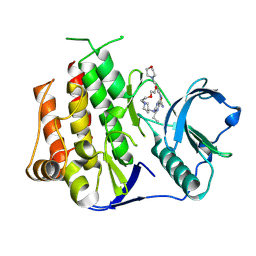 | | Crystal structure of FLT3 in complex with a Pyrazinamide Macrocycle derivative | | Descriptor: | 3^3-ethyl-5^4-morpholino-6,9-dioxa-2,4-diaza-3(2,6)-pyrazina-1(4,1)-piperidina-5(1,3)-benzenacycloundecaphane-3^5-carboxamide, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Guo, M, Chen, Y. | | Deposit date: | 2023-12-05 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Based Optimization of Pyrazinamide-Containing Macrocyclic Derivatives as Fms-like Tyrosine Kinase 3 (FLT3) Inhibitors to Overcome Clinical Mutations.
Acs Pharmacol Transl Sci, 7, 2024
|
|
2PIG
 
 | | Crystal structure of ydcK from Salmonella cholerae at 2.38 A resolution. Northeast Structural Genomics target SCR6 | | Descriptor: | Putative transferase, ZINC ION | | Authors: | Benach, J, Chen, Y, Vorobiev, S.M, Seetharaman, J, Ho, C.K, Janjua, H, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ydcK from Salmonella cholerae at 2.38 A resolution.
To be Published
|
|
2PX9
 
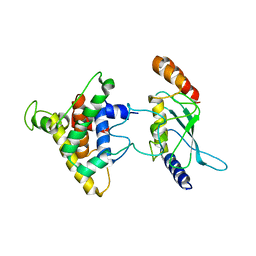 | | The intrinsic affinity between E2 and the Cys domain of E1 in Ubiquitin-like modifications | | Descriptor: | SUMO-activating enzyme subunit 2, SUMO-conjugating enzyme UBC9 | | Authors: | Wang, J.H, Hu, W.D, Cai, S, Lee, B, Song, J, Chen, Y. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications.
Mol.Cell, 27, 2007
|
|
6JPE
 
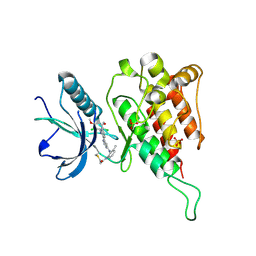 | | Crystal structure of FGFR4 kinase domain with irreversible inhibitor 1 | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[6-[2-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]amino]pyridin-3-yl]pyrimidin-4-yl]amino]-3-methyl-phenyl]prop-2-enamide, SULFATE ION | | Authors: | Chen, X, Dai, S, Zhou, Z, Chen, Y. | | Deposit date: | 2019-03-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Development of a Potent and Specific FGFR4 Inhibitor for the Treatment of Hepatocellular Carcinoma.
J.Med.Chem., 63, 2020
|
|
6JBH
 
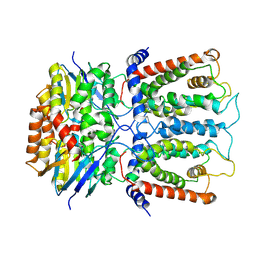 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
8FTL
 
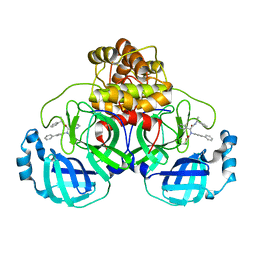 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1
To Be Published
|
|
4E0R
 
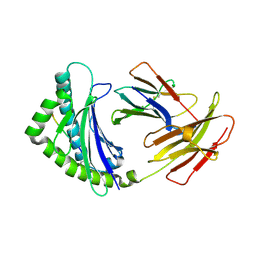 | | Structure of the chicken MHC class I molecule BF2*0401 | | Descriptor: | 8-MERIC PEPTIDE (FUS/TLS), Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
2O3I
 
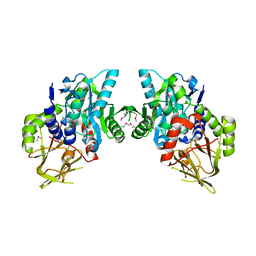 | | X-ray Crystal Structure of Protein CV_3147 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR68. | | Descriptor: | Hypothetical protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Cunningham, K, Ma, L.C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the hypothetical protein Q7NTB2_CHRVO from Chromobacterium violaceum
To be Published
|
|
4G42
 
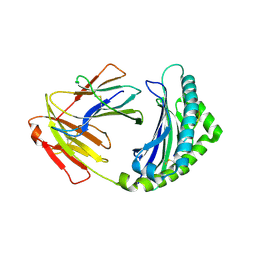 | | Structure of the Chicken MHC Class I Molecule BF2*0401 complexed to pepitde P8D | | Descriptor: | 8-MERIC PEPTIDE P8D, Beta-2 microglobulin, MHC class I alpha chain 2 | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
8HN6
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HN7
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
6M23
 
 | | Overall structure of KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chi, X.M, Li, X.R, Chen, Y, Zhang, Y.Y, Su, Q, Zhou, Q. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the full-length human KCC2 and KCC3 cation-chloride cotransporters.
Cell Res., 31, 2021
|
|
2QYK
 
 | | Crystal structure of PDE4A10 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, Cyclic AMP-specific phosphodiesterase HSPDE4A10, MAGNESIUM ION, ... | | Authors: | Wang, H, Peng, M, Chen, Y, Geng, J, Robinson, H, Houslay, M. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
4G43
 
 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
7K99
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 | | Descriptor: | (hydroxy{(1S)-1-(methylsulfanyl)-2-[5-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)-1H-benzotriazol-1-yl]ethyl}amino)methanol, GLYCEROL, SULFATE ION, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
2P0V
 
 | | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 | | Descriptor: | Hypothetical protein BT3781 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
