4RNU
 
 | | G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RP6
 
 | |
6MZM
 
 | | Human TFIID bound to promoter DNA and TFIIA | | Descriptor: | SCP DNA (80-MER), TATA-box-binding protein, Transcription initiation factor IIA subunit 1, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6NLK
 
 | | 1.85 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 13) | | Descriptor: | 4-{[3-(4-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NLG
 
 | | 1.50 A resolution structure of BfrB (C89S/K96C) from Pseudomonas aeruginosa in complex with a small molecule fragment (analog 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-hydroxy-1H-isoindole-1,3(2H)-dione, Bacterioferritin, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NUT
 
 | | Ebola virus nucleoprotein - RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3') | | Authors: | Kirchdoerfer, R.N, Ward, A.B. | | Deposit date: | 2019-02-01 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4NCO
 
 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP gp120, BG505 SOSIP gp41, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Lyumkis, D, Ward, A.B, Wilson, I.A. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Crystal structure of a soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
4RNV
 
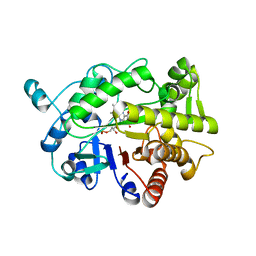 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4O3Y
 
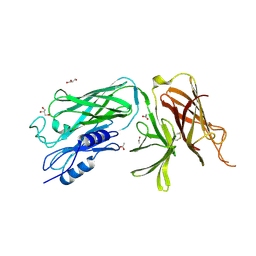 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Arg-179-Glu from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O4X
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) double mutant Tyr-167-Ala and Trp-176-Ala from Haemophilus parasuis Hp5 | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4RNW
 
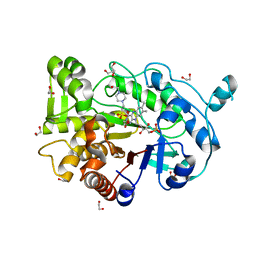 | | Truncated version of the G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4O3X
 
 | |
4TLW
 
 | | CARDS TOXIN, FULL-LENGTH | | Descriptor: | ADP-ribosylating toxin CARDS | | Authors: | Becker, A, GALALELDEEN, A, Taylor, A.B, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4O3W
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-63-Ala from Actinobacillus suis H57 | | Descriptor: | GLYCEROL, SULFATE ION, Transferrin binding protein B | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O4U
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Trp-176-Ala from Haemophilus parasuis Hp5 | | Descriptor: | GLYCEROL, TbpB | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4O49
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Tyr-174-Ala from Actinobacillus pleuropneumoniae H87 | | Descriptor: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | Authors: | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | Deposit date: | 2013-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
4TKP
 
 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-05-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
4TLV
 
 | | CARDS TOXIN, NICKED | | Descriptor: | ACETATE ION, ADP-ribosylating toxin CARDS, GLYCEROL, ... | | Authors: | Taylor, A.B, Pakhomova, O.N, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RNX
 
 | | K154 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1 | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
3FOU
 
 | |
3DDU
 
 | | Prolyl Oligopeptidase with GSK552 | | Descriptor: | (6S)-1-chloro-3-[(4-fluorobenzyl)oxy]-6-(pyrrolidin-1-ylcarbonyl)pyrrolo[1,2-a]pyrazin-4(6H)-one, ACETATE ION, GLYCEROL, ... | | Authors: | Madauss, K.P, Reid, R.A, Haffner, C.D, Miller, A.B. | | Deposit date: | 2008-06-06 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Pyrrolidinyl pyridone and pyrazinone analogues as potent inhibitors of prolyl oligopeptidase (POP)
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CQP
 
 | | Human SOD1 G85R Variant, Structure I | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
3DT3
 
 | | Human Estrogen receptor alpha LBD with GW368 | | Descriptor: | 5-(4-hydroxyphenoxy)-6-(3-hydroxyphenyl)-7-methylnaphthalen-2-ol, Estrogen receptor | | Authors: | Williams, S.P, Miller, A.B. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of 3-alkyl naphthalenes as novel estrogen receptor ligands.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GZO
 
 | | HUMAN SOD1 G93A Variant | | Descriptor: | COPPER (II) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Galaleldeen, A, Taylor, A.B, Narayana, N, Whitson, L.J, Hart, P.J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
3H2Q
 
 | | Human SOD1 H80R variant, P21 crystal form | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Seetharaman, S.V, Winkler, D.D, Taylor, A.B, Cao, X, Whitson, L.J, Doucette, P.A, Valentine, J.S, Carroll, M.C, Culotta, V.C, Hart, P.J. | | Deposit date: | 2009-04-14 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of Pathogenic SOD1 Mutants H80R and D124V: Disrupted Zinc-binding
and Compromised Post-translational Modification by the Copper Chaperone CCS
To be Published
|
|
