6HP8
 
 | | Crystal structure of DPP8 in complex with Val-BoroPro | | Descriptor: | Dipeptidyl peptidase 8, GLYCEROL, SODIUM ION, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Improvement of Protein Crystal Diffraction Using Post-Crystallization Methods: Infrared Laser Radiation Controls Crystal Order
Thesis, 2019
|
|
1BKC
 
 | | CATALYTIC DOMAIN OF TNF-ALPHA CONVERTING ENZYME (TACE) | | Descriptor: | N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TUMOR NECROSIS FACTOR-ALPHA-CONVERTING ENZYME, ZINC ION | | Authors: | Maskos, K, Fernandez-Catalan, C, Bode, W. | | Deposit date: | 1998-04-23 | | Release date: | 1999-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3ZI5
 
 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | Descriptor: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | Authors: | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2013-01-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
4AIG
 
 | | ADAMALYSIN II WITH PHOSPHONATE INHIBITOR | | Descriptor: | ADAMALYSIN II, CALCIUM ION, N-[(FURAN-2-YL)CARBONYL]-(S)-LEUCYL-(R)-[1-AMINO-2(1H-INDOL-3-YL)ETHYL]-PHOSPHONIC ACID, ... | | Authors: | Pochetti, G, Mazza, F, Gavuzzo, E, Cirilli, M. | | Deposit date: | 1997-10-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2 angstrom X-ray structure of adamalysin II complexed with a peptide phosphonate inhibitor adopting a retro-binding mode.
FEBS Lett., 418, 1997
|
|
1AI8
 
 | | HUMAN ALPHA-THROMBIN TERNARY COMPLEX WITH THE EXOSITE INHIBITOR HIRUGEN AND ACTIVE SITE INHIBITOR PHCH2OCO-D-DPA-PRO-BOROMPG | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN IIIB, ... | | Authors: | Skordalakes, E, Dodson, G, Elgendy, S, Goodwin, C.A, Green, D, Tyrrel, R, Scully, M.F, Freyssinet, J, Kakkar, V.V, Deadman, J. | | Deposit date: | 1997-05-01 | | Release date: | 1997-10-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships.
Protein Sci., 1, 1992
|
|
4BFH
 
 | |
4THN
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-THROMBIN-HIRUNORM IV COMPLEX REVEALS A NOVEL SPECIFICITY SITE RECOGNITION MODE. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUNORM IV | | Authors: | Lombardi, A, De Simone, G, Nastri, F, Galdiero, S, Della Morte, R, Staiano, N, Pedone, C, Bolognesi, M, Pavone, V. | | Deposit date: | 1998-09-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of alpha-thrombin-hirunorm IV complex reveals a novel specificity site recognition mode.
Protein Sci., 8, 1999
|
|
4PGT
 
 | | CRYSTAL STRUCTURE OF HGSTP1-1[V104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Blaszczyk, J. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
1AIN
 
 | |
1A4A
 
 | |
1AMZ
 
 | |
1AL6
 
 | |
1A4B
 
 | |
1A85
 
 | | MMP8 WITH MALONIC AND ASPARAGINE BASED INHIBITOR | | Descriptor: | CALCIUM ION, MMP-8, N~1~-(3-aminobenzyl)-N~2~-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-aspartamide, ... | | Authors: | Brandstetter, H, Roedern, E.G.V, Grams, F, Engh, R.A. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of malonic acid-based inhibitors bound to human neutrophil collagenase. A new binding mode explains apparently anomalous data.
Protein Sci., 7, 1998
|
|
1A4C
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY HIS, PH 3.5 CRYSTAL FORM, DATA COLLECTED AT-180 DEGREES CELSIUS | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION, ... | | Authors: | Messerschmidt, A, Prade, L. | | Deposit date: | 1998-01-28 | | Release date: | 1998-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Rack-induced metal binding vs. flexibility: Met121His azurin crystal structures at different pH.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1ABI
 
 | |
1A2C
 
 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
1A86
 
 | | MMP8 WITH MALONIC AND ASPARTIC ACID BASED INHIBITOR | | Descriptor: | CALCIUM ION, MMP-8, N-benzyl-N~2~-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-alpha-asparagine, ... | | Authors: | Brandstetter, H, Roedern, E.G.V, Grams, F, Engh, R.A. | | Deposit date: | 1998-04-03 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of malonic acid-based inhibitors bound to human neutrophil collagenase. A new binding mode explains apparently anomalous data.
Protein Sci., 7, 1998
|
|
1ABJ
 
 | |
1AYE
 
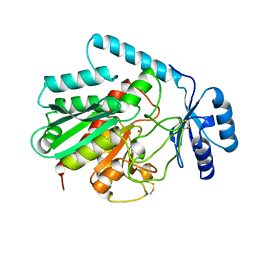 | | HUMAN PROCARBOXYPEPTIDASE A2 | | Descriptor: | PROCARBOXYPEPTIDASE A2, ZINC ION | | Authors: | Garcia-Saez, I, Reverte, D, Vendrell, J, Aviles, F.X, Coll, M. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of human procarboxypeptidase A2. Deciphering the basis of the inhibition, activation and intrinsic activity of the zymogen.
EMBO J., 16, 1997
|
|
1BBR
 
 | |
1BQY
 
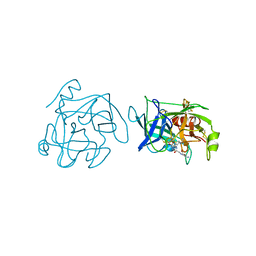 | | Plasminogen activator (TSV-PA) from snake venom | | Descriptor: | L-alpha-glutamyl-N-{(1S)-4-{[amino(iminio)methyl]amino}-1-[(1S)-2-chloro-1-hydroxyethyl]butyl}glycinamide, PLASMINOGEN ACTIVATOR | | Authors: | Parry, M.A.A, Bode, W. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the novel snake venom plasminogen activator TSV-PA: a prototype structure for snake venom serine proteinases.
Structure, 6, 1998
|
|
1BQB
 
 | | AUREOLYSIN, STAPHYLOCOCCUS AUREUS METALLOPROTEINASE | | Descriptor: | CALCIUM ION, PROTEIN (AUREOLYSIN), ZINC ION | | Authors: | Medrano, F.J, Banbula, A, Potempa, J, Travis, J, Bode, W. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72 A resolution.
Structure, 6, 1998
|
|
2OVO
 
 | |
3HGS
 
 | | Crystal structure of tomato OPR3 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
