8SGW
 
 | | Pendrin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SHC
 
 | | Pendrin in complex with Niflumic acid | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CHLORIDE ION, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SIE
 
 | | Pendrin in complex with bicarbonate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, BICARBONATE ION, CHOLESTEROL, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
4YOQ
 
 | | Crystal Structure of MutY bound to its anti-substrate | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(*T*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJV
 
 | | Crystal structure of human granzyme H with a peptidyl substrate | | Descriptor: | Granzyme H, PTSYAGDDSG, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
1BLR
 
 | | NMR SOLUTION STRUCTURE OF HUMAN CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II, 22 STRUCTURES | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II | | Authors: | Wang, L, Li, Y, Abilddard, F, Yan, H, Markely, J. | | Deposit date: | 1998-07-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of type II human cellular retinoic acid binding protein: implications for ligand binding.
Biochemistry, 37, 1998
|
|
8WWC
 
 | |
6BFN
 
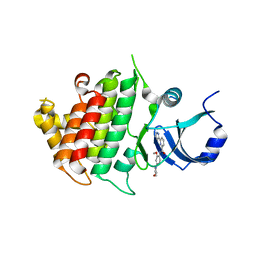 | | Crystal structure of human IRAK1 | | Descriptor: | Interleukin-1 receptor-associated kinase 1, N-[2-methoxy-4-(morpholin-4-yl)phenyl]-6-(1H-pyrazol-5-yl)pyridine-2-carboxamide | | Authors: | Wang, L, Qiao, Q, Wu, H. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human IRAK1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6D26
 
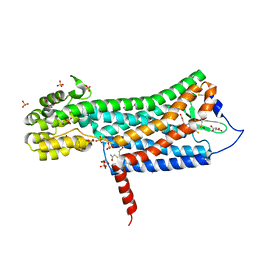 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6D27
 
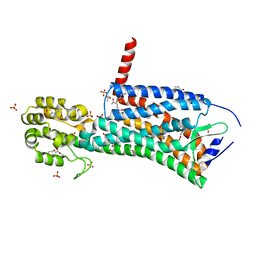 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6EGA
 
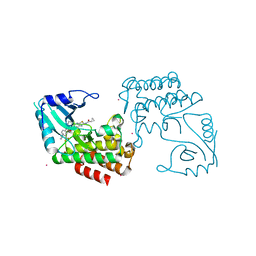 | | IRAK4 in complex with a type II inhibitor | | Descriptor: | 3-{2-[(cyclopropanecarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(piperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
7Y0O
 
 | |
7Y0N
 
 | | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R15-F7, ... | | Authors: | Wang, L. | | Deposit date: | 2022-06-05 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 WT Spike in complex with R15 Fab and P14 Nanobody
To Be Published
|
|
7VHM
 
 | |
7YR4
 
 | |
5YR2
 
 | | Structure of cpGFP66BPA | | Descriptor: | Green fluorescent protein | | Authors: | Wang, L, Kang, F, Wang, J. | | Deposit date: | 2017-11-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure of cpGFP66BPA
To Be Published
|
|
7E1A
 
 | |
7ZYU
 
 | |
7DV5
 
 | |
2LKU
 
 | |
2LKT
 
 | |
8UUK
 
 | | Pendrin in apo | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
7UPT
 
 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (open conformation) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
