3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VF5
 
 | | Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3CYX
 
 | | Crystal structure of HIV-1 mutant I50V and inhibitor saquinavira | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, ACETIC ACID, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3CYW
 
 | | Effect of Flap Mutations on Structure of HIV-1 Protease and Inhibition by Saquinavir and Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D1X
 
 | | Crystal structure of HIV-1 mutant I54M and inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D1Y
 
 | | Crystal structure of HIV-1 mutant I54V and inhibitor SAQUINA | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
2YN5
 
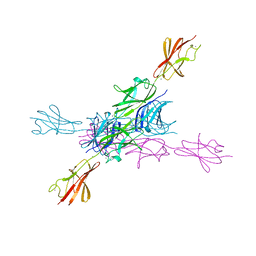 | | Structural insight into the giant calcium-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells | | Descriptor: | CALCIUM ION, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Griessl, M.H, Schmid, B, Kassler, K, Braunsmann, C, Ritter, R, Barlag, B, Sturm, K.U, Danzer, C, Wagner, C, Schaeffer, T.E, Sticht, H, Hensel, M, Muller, Y.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insight Into the Giant Ca(2+)-Binding Adhesin Siie: Implications for the Adhesion of Salmonella Enterica to Polarized Epithelial Cells.
Structure, 21, 2013
|
|
2YN3
 
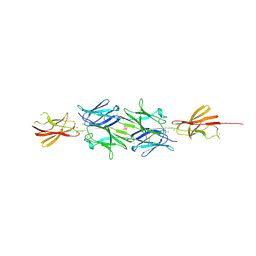 | | Structural insight into the giant calcium-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells | | Descriptor: | CALCIUM ION, IODIDE ION, PHOSPHATE ION, ... | | Authors: | Griessl, M.H, Schmid, B, Kassler, K, Braunsmann, C, Ritter, R, Barlag, B, Sturm, K.U, Danzer, C, Wagner, C, Schaeffer, T.E, Sticht, H, Hensel, M, Muller, Y.A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Insight Into the Giant Ca(2+)-Binding Adhesin Siie: Implications for the Adhesion of Salmonella Enterica to Polarized Epithelial Cells.
Structure, 21, 2013
|
|
5IQ5
 
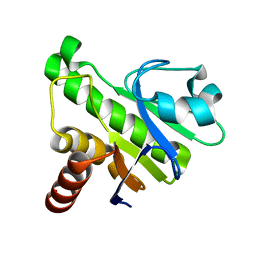 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
2VT0
 
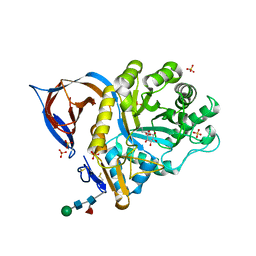 | | X-ray structure of a conjugate with conduritol-beta-epoxide of acid-beta-glucosidase overexpressed in cultured plant cells | | Descriptor: | (1R,2R,3S,4S,5S,6S)-CYCLOHEXANE-1,2,3,4,5,6-HEXOL, GLUCOSYLCERAMIDASE, SULFATE ION, ... | | Authors: | Brumshtein, B, Greenblatt, H.M, Shaaltiel, Y, Aviezer, D, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2008-05-03 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Acid Beta-Glucosidase: Insights from Structural Analysis and Relevance to Gaucher Disease Therapy.
Biol.Chem., 389, 2008
|
|
1YZB
 
 | | Solution structure of the Josephin domain of Ataxin-3 | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Nicastro, G, Masino, L, Menon, R.P, Knowles, P.P, McDonald, N.Q, Pastore, A. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Josephin domain of ataxin-3: Structural determinants for molecular recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4RAV
 
 | | Crystal structure of scFvC4 in complex with the first 17 AA of huntingtin | | Descriptor: | Huntingtin, SULFATE ION, Single-chain Fv, ... | | Authors: | De Genst, E, Chirgadze, D.Y, Dobson, C.M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a single-chain fv bound to the 17 N-terminal residues of huntingtin provides insights into pathogenic amyloid formation and suppression.
J.Mol.Biol., 427, 2015
|
|
4BRX
 
 | | Focal Adhesion Kinase catalytic domain in complex with a diarylamino- 1,3,5-triazine inhibitor | | Descriptor: | 2-(4-(2-methoxy-4-morpholinophenylamino)-1,3,5-triazin-2-ylamino)-N-methylbenzamide, FOCAL ADHESION KINASE 1, SULFATE ION | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of Novel Diarylamino-1,3,5-Triazine Derivatives as Fak Inhibitors with Anti-Angiogenic Activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4C7T
 
 | | Focal Adhesion Kinase catalytic domain in complex with a diarylamino- 1,3,5-triazine inhibitor | | Descriptor: | FOCAL ADHESION KINASE 1, N-methyl-2-[[4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl]amino]benzamide, SULFATE ION | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2013-09-25 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Inhibition of Both Focal Adhesion Kinase and Fibroblast Growth Factor Receptor 2 Pathways Induces Anti-Tumor and Anti-Angiogenic Activities.
Cancer Lett., 348, 2014
|
|
7EZX
 
 | |
