3PUP
 
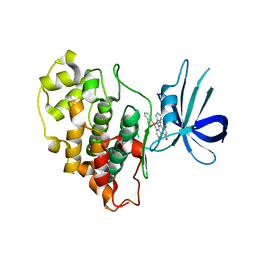 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
5JRQ
 
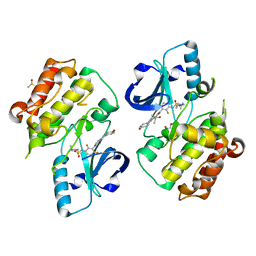 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-6-VEM | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{2,4-difluoro-3-[5-(4-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]phenyl}propane-1-sulfonamide, ... | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
5JSM
 
 | |
5U2J
 
 | |
3ZQD
 
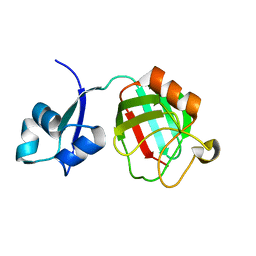 | | B. subtilis L,D-transpeptidase | | Descriptor: | L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J.-P, Bougault, C, Arthur, M, Hugonnet, J.-E, Veckerle, C, Pessey, O. | | Deposit date: | 2011-06-09 | | Release date: | 2012-05-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
4WJY
 
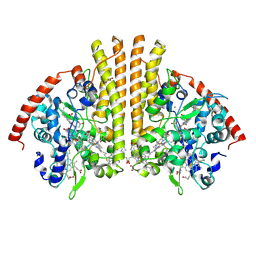 | | Esherichia coli nitrite reductase NrfA H264N | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Edwards, M.J, Lockwood, C.W.J. | | Deposit date: | 2014-10-01 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Resolution of key roles for the distal pocket histidine in cytochrome C nitrite reductases.
J.Am.Chem.Soc., 137, 2015
|
|
6PGQ
 
 | |
4A52
 
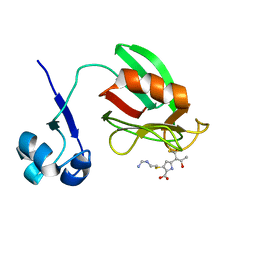 | | NMR structure of the imipenem-acylated L,D-transpeptidase from Bacillus subtilis | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, PUTATIVE L, D-TRANSPEPTIDASE YKUD | | Authors: | Lecoq, L, Simorre, J, Bougault, C, Arthur, M, Hugonnet, J, Veckerle, C, Pessey, O. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-30 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Dynamics Induced by Beta-Lactam Antibiotics in the Active Site of Bacillus subtilis L,D-Transpeptidase.
Structure, 20, 2012
|
|
7KQD
 
 | | Prefusion RSV F Bound to RV521 | | Descriptor: | 1'-{[5-(aminomethyl)-1-(4,4,4-trifluorobutyl)-1H-benzimidazol-2-yl]methyl}-6'-fluorospiro[cyclopropane-1,3'-indol]-2'(1'H)-one, Fusion glycoprotein F0, SULFATE ION | | Authors: | McLellan, J.S. | | Deposit date: | 2020-11-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Discovery of Sisunatovir (RV521), an Inhibitor of Respiratory Syncytial Virus Fusion.
J.Med.Chem., 64, 2021
|
|
7UOQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH (2S)-2-(TERT-BUTOXY)-2-(5-{2-[(2-CHLORO-6-M ETHYLPHENYL)METHYL]-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL}-4- (4,4-DIMETHYLPIPERIDIN-1-YL)-2-METHYLPYRIDIN-3-YL)ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(5M)-5-{2-[(2-chloro-6-methylphenyl)methyl]-1,2,3,4-tetrahydroisoquinolin-6-yl}-4-(4,4-dimethylpiperidin-1-yl)-2-methylpyridin-3-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-04-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8867 Å) | | Cite: | Discovery and Preclinical Profiling of GSK3839919, a Potent HIV-1 Allosteric Integrase Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
5FCM
 
 | | CrBld10-N 1-70 | | Descriptor: | Basal body protein, ISOPROPYL ALCOHOL, SULFATE ION | | Authors: | Kraatz, S.H.W, Steinmetz, M.O. | | Deposit date: | 2015-12-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | The Human Centriolar Protein CEP135 Contains a Two-Stranded Coiled-Coil Domain Critical for Microtubule Binding.
Structure, 24, 2016
|
|
5FCN
 
 | | microtubule binding domain of human CEP135 | | Descriptor: | Centrosomal protein of 135 kDa | | Authors: | Kraatz, S.H.W. | | Deposit date: | 2015-12-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Human Centriolar Protein CEP135 Contains a Two-Stranded Coiled-Coil Domain Critical for Microtubule Binding.
Structure, 24, 2016
|
|
7U2U
 
 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH Compound-2a AKA (2S)-2-(TERT-BUTOXY)-2-[7-(4,4-DIMETHYLPIPE RIDIN-1-YL)-8-{4-[2-(4-FLUOROPHENYL)ETHOXY]PHENYL}-2,5-DIM ETHYLIMIDAZO[1,2-A]PYRIDIN-6-YL]ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(4S)-7-(4,4-dimethylpiperidin-1-yl)-8-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,5-dimethylimidazo[1,2-a]pyridin-6-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Khan, J.A, lewis, H, Kish, K. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Scaffold modifications to the 4-(4,4-dimethylpiperidinyl) 2,6-dimethylpyridinyl class of HIV-1 allosteric integrase inhibitors.
Bioorg.Med.Chem., 67, 2022
|
|
2BS3
 
 | |
2BS4
 
 | | GLU C180 -> ILE VARIANT QUINOL:FUMARATE REDUCTASE FROMWOLINELLA SUCCINOGENES | | Descriptor: | 2,3-DIMETHYL-1,4-NAPHTHOQUINONE, CITRIC ACID, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Experimental Support for the E-Pathway Hypothesis of Coupled Transmembrane Electron and Proton Transfer in Dihemic Quinol:Fumarate Reductase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8FIU
 
 | | Potent long-acting inhibitors targeting HIV-1 capsid based on a versatile quinazolin-4-one scaffold | | Descriptor: | 1,2-ETHANEDIOL, HIV-1 capsid, N-[(1S)-1-{(3P)-3-{4-chloro-3-[(methanesulfonyl)amino]-1-methyl-1H-indazol-7-yl}-7-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-oxo-3,4-dihydroquinazolin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Nolte, R.T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
J.Med.Chem., 66, 2023
|
|
2BS2
 
 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Evidence for Transmembrane Proton Transfer in a Dihaem-Containing Membrane Protein Complex.
Embo J., 25, 2006
|
|
5VAR
 
 | |
3RAE
 
 | | Quinolone(Levofloxacin)-DNA cleavage complex of type IV topoisomerase from S. pneumoniae | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, 5'-D(*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*GP*TP*GP*CP*AP*T)-3', ... | | Authors: | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2011-03-28 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3BNG
 
 | | W. succinogenes NrfA Y218F | | Descriptor: | ACETATE ION, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Lukat, P, Einsle, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding and Reduction of Sulfite by Cytochrome c Nitrite Reductase
Biochemistry, 47, 2008
|
|
3BNJ
 
 | | W. succinogenes NrfA Y218F Sulfite Complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Lukat, P, Einsle, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding and Reduction of Sulfite by Cytochrome c Nitrite Reductase
Biochemistry, 47, 2008
|
|
3BNF
 
 | | W. succinogenes NrfA Sulfite Complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Lukat, P, Einsle, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and Reduction of Sulfite by Cytochrome c Nitrite Reductase
Biochemistry, 47, 2008
|
|
3BNH
 
 | | W. succinogenes NrfA Y218F Nitrite Complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Lukat, P, Einsle, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding and Reduction of Sulfite by Cytochrome c Nitrite Reductase
Biochemistry, 47, 2008
|
|
1E7P
 
 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D, Kroeger, A. | | Deposit date: | 2000-09-01 | | Release date: | 2001-04-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Third Crystal Form of Wolinella Succinogenes Quinol:Fumarate Reductase Reveals Domain Closure at the Site of Fumarate Reduction
Eur.J.Biochem., 268, 2001
|
|
2M32
 
 | | Alpha-1 integrin I-domain in complex with GLOGEN triple helical peptide | | Descriptor: | GLOGEN peptide, Integrin alpha-1, MAGNESIUM ION | | Authors: | Chin, Y, Headey, S, Mohanty, B, McEwan, P, Swarbrick, J, Mulhern, T, Emsley, J, Simpson, J, Scanlon, M. | | Deposit date: | 2013-01-07 | | Release date: | 2013-11-06 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | The Structure of Integrin alpha 1I Domain in Complex with a Collagen-mimetic Peptide.
J.Biol.Chem., 288, 2013
|
|
