4TTT
 
 | |
5MDJ
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in a its as-isolated high-pressurized form | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G1Z
 
 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its Pfr state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Sauthof, L, Schmidt, A, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
8POX
 
 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.6 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
8POW
 
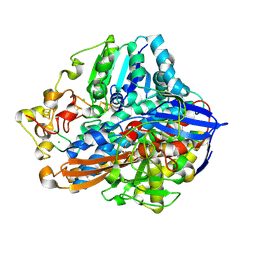 | | Crystal Structure of the C19G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the air-oxidized state at 1.61 A Resolution. | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Fe4S4, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
3BB0
 
 | |
3AZU
 
 | |
3G7K
 
 | |
3GK7
 
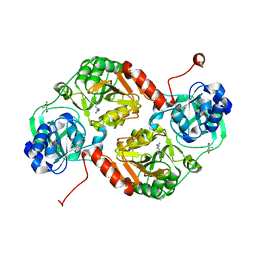 | | Crystal structure of 4-hydroxybutyrate CoA-Transferase from Clostridium aminobutyricum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-Hydroxybutyrate CoA-transferase, SPERMIDINE | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M. | | Deposit date: | 2009-03-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum
Biol.Chem., 390, 2009
|
|
3QDQ
 
 | |
5L6Q
 
 | | Refolded AL protein from cardiac amyloidosis | | Descriptor: | CARBONATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Annamalai, K, Liberta, F, Vielberg, M.-T, Lilie, H, Guehrs, K.-H, Schierhorn, A, Koehler, R, Schmidt, A, Haupt, C, Hegenbart, O, Schoenland, S, Groll, M, Faendrich, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Common Fibril Structures Imply Systemically Conserved Protein Misfolding Pathways In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2W4R
 
 | | Crystal structure of the regulatory domain of human LGP2 | | Descriptor: | MERCURY (II) ION, PROBABLE ATP-DEPENDENT RNA HELICASE DHX58, SULFATE ION | | Authors: | Pippig, D.A, Hellmuth, J.C, Cui, S, Kirchhofer, A, Lammens, K, Lammens, A, Schmidt, A, Rothenfusser, S, Hopfner, K.P. | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Regulatory Domain of the Rig-I Family ATPase Lgp2 Senses Double-Stranded RNA.
Nucleic Acids Res., 37, 2009
|
|
7PIU
 
 | | Cryo-EM structure of the agonist setmelanotide bound to the active melanocortin-4 receptor (MC4R) in complex with the heterotrimeric Gs protein at 2.6 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
7PIV
 
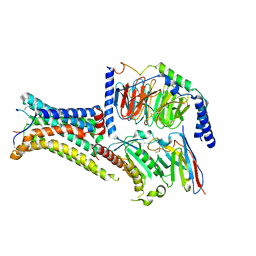 | | Active Melanocortin-4 receptor (MC4R)- Gs protein complex bound to agonist NDP-alpha-MSH at 2.86 A resolution. | | Descriptor: | CALCIUM ION, Camelid antibody VHH fragment - nanobody 35, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Heyder, N.A, Schmidt, A, Kleinau, G, Hilal, T, Scheerer, P. | | Deposit date: | 2021-08-23 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Cell Res., 31, 2021
|
|
5T4X
 
 | | CRYSTAL STRUCTURE OF PDE6D IN APO-STATE | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Qureshi, B.M, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-08-30 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanistic insights into the role of prenyl-binding protein PrBP/ delta in membrane dissociation of phosphodiesterase 6.
Nat Commun, 9, 2018
|
|
2B97
 
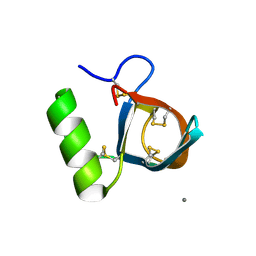 | | Ultra-high resolution structure of hydrophobin HFBII | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Linder, M, Popov, A, Schmidt, A, Rouvinen, J. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5FLX
 
 | | Mammalian 40S HCV-IRES complex | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yamamoto, H, Collier, M, Loerke, J, Ismer, J, Schmidt, A, Hilal, T, Sprink, T, Yamamoto, K, Mielke, T, Burger, J, Shaikh, T.R, Dabrowski, M, Hildebrand, P.W, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Architecture of the Ribosome-Bound Hepatitis C Virus Internal Ribosomal Entry Site RNA.
Embo J., 34, 2015
|
|
1IBJ
 
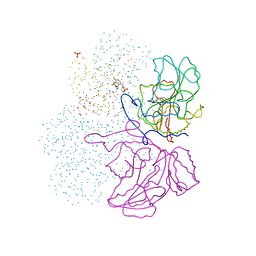 | | Crystal structure of cystathionine beta-lyase from Arabidopsis thaliana | | Descriptor: | CARBONATE ION, CYSTATHIONINE BETA-LYASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Breitinger, U, Clausen, T, Messerschmidt, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity
Plant Physiol., 126, 2001
|
|
1BWW
 
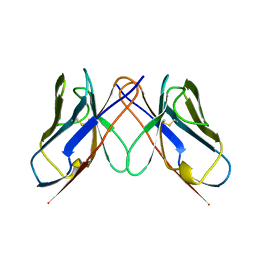 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | Descriptor: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | Authors: | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6GYH
 
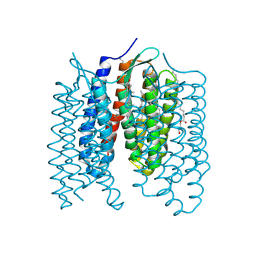 | | Crystal structure of the light-driven proton pump Coccomyxa subellipsoidea Rhodopsin CsR | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Family A G protein-coupled receptor-like protein, ... | | Authors: | Szczepek, M, Schmidt, A, Scheerer, P. | | Deposit date: | 2018-06-29 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a light-gated proton channel based on the crystal structure ofCoccomyxarhodopsin.
Sci.Signal., 12, 2019
|
|
8AZW
 
 | | Cryo-EM structure of the plant 60S subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8AUV
 
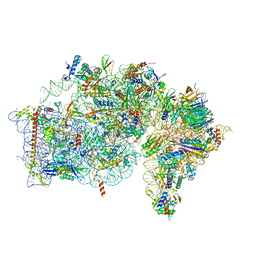 | | Cryo-EM structure of the plant 40S subunit | | Descriptor: | 18S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
5MDL
 
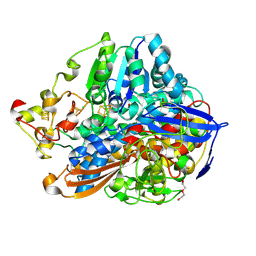 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its O2-derivatized form by a "soak-and-freeze" derivatization method | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Kalms, J, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8B2L
 
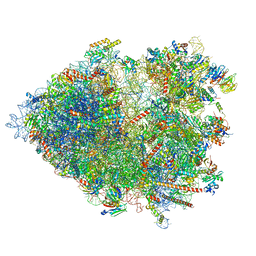 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
2FSS
 
 | |
