7ZU2
 
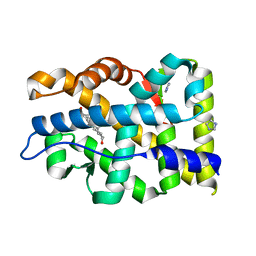 | | Crystal structure of mutant AR-LBD (Q799E) bound to dihydrotestosterone | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, IMIDAZOLE | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
7ZU1
 
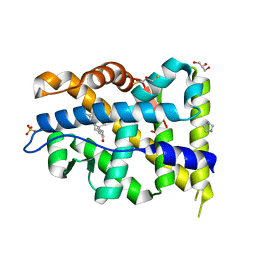 | | Crystal structure of mutant AR-LBD (V758A) bound to dihydrotestosterone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Alegre-Marti, A, Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-05-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance.
Sci Adv, 9, 2023
|
|
8RKF
 
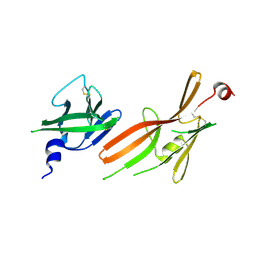 | | Crystal structure of the ZP-N1 and ZP-N2 domains of human ZP2 (hZP2-N1N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Stsiapanava, A, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
3RJA
 
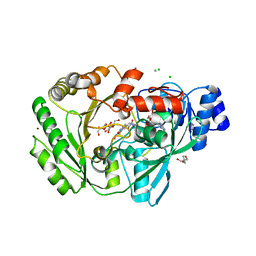 | | Crystal structure of carbohydrate oxidase from Microdochium nivale in complex with substrate analogue | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duskova, J, Skalova, T, Kolenko, P, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
3RJ8
 
 | | Crystal structure of carbohydrate oxidase from Microdochium nivale | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carbohydrate oxidase, ... | | Authors: | Duskova, J, Skalova, T, Stepankova, A, Koval, T, Hasek, J, Ostergaard, L.H, Fuglsang, C.C, Kolenko, P, Dohnalek, J. | | Deposit date: | 2011-04-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and kinetic studies of carbohydrate oxidase from Microdochium nivale
To be Published
|
|
3SK0
 
 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2011-06-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
7YXP
 
 | | Crystal structure of WT AncGR2-LBD WT bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXR
 
 | | Crystal structure of mutant AncGR2-LBD (Y545A) bound to dexamethasone and SHP coregulator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, FORMIC ACID, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXO
 
 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7YXN
 
 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, FORMIC ACID, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
7SIU
 
 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
5WBE
 
 | | COX-1:MOFEZOLAC COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mofezolac, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
5FTQ
 
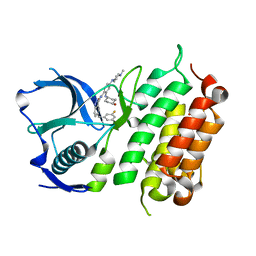 | | Crystal structure of the ALK kinase domain in complex with Cmpd 17 | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, GLYCEROL, N-[5-(3,5-DIFLUOROBENZYL)-1H-INDAZOL-3-YL]-2-[(4-HYDROXYCYCLOHEXYL)AMINO]-4-(4-METHYLPIPERAZIN-1-YL) BENZAMIDE | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
1I5I
 
 | | THE C18S MUTANT OF BOVINE (GAMMA-B)-CRYSTALLIN | | Descriptor: | (GAMMA-B) CRYSTALLIN | | Authors: | Zarutskie, J.A, Asherie, N, Pande, J, Pande, A, Lomakin, J, Lomakin, A, Ogun, O, Stern, L.J, King, J.A, Benedek, G.B. | | Deposit date: | 2001-02-27 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enhanced crystallization of the Cys18 to Ser mutant of bovine gammaB crystallin.
J.Mol.Biol., 314, 2001
|
|
5FTO
 
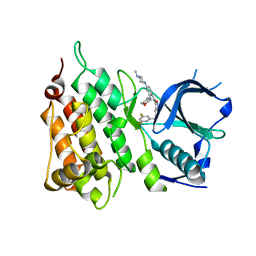 | | Crystal structure of the ALK kinase domain in complex with Entrectinib | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, Entrectinib | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
4KMG
 
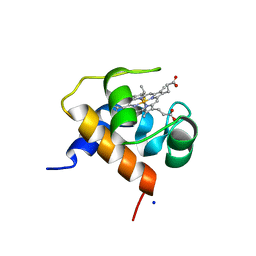 | | Crystal structure of cytochrome c6B from Synechococcus sp. WH8102 | | Descriptor: | Cytochrome C6 (Soluble cytochrome F) (Cytochrome c553), HEME C, SODIUM ION | | Authors: | Zatwarnicki, P, Krzywda, S, Barciszewski, J, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2013-05-08 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome c6B of Synechococcus sp. WH 8102 - Crystal structure and basic properties of novel c6-like family representative.
Biochem.Biophys.Res.Commun., 443, 2014
|
|
3JB4
 
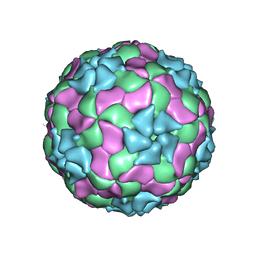 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
2A4R
 
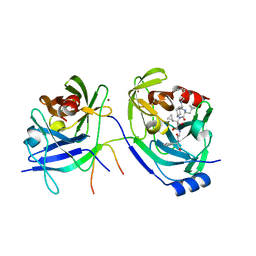 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3Q20
 
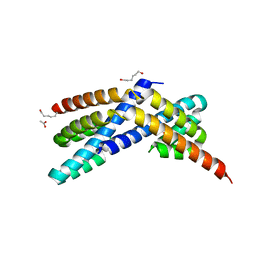 | | Crystal structure of RbcX C103A mutant from Thermosynechococcus elongatus | | Descriptor: | ACETATE ION, HEXANE-1,6-DIOL, RbcX protein | | Authors: | Tarnawski, M, Krzywda, S, Szczepaniak, A, Jaskolski, M. | | Deposit date: | 2010-12-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of the RuBisCO chaperone RbcX from the thermophilic cyanobacterium Thermosynechococcus elongatus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3DR0
 
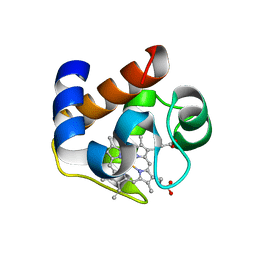 | | Structure of reduced cytochrome c6 from Synechococcus sp. PCC 7002 | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Bialek, W, Krzywda, S, Jaskolski, M, Szczepaniak, A. | | Deposit date: | 2008-07-10 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic-resolution structure of reduced cyanobacterial cytochrome c6 with an unusual sequence insertion
Febs J., 276, 2009
|
|
3RK4
 
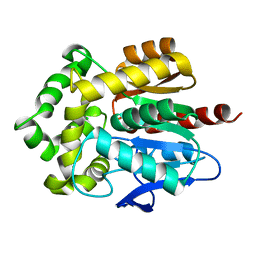 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3SNG
 
 | | X-ray structure of fully glycosylated bifunctional nuclease TBN1 from Solanum lycopersicum (Tomato) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koval, T, Stepankova, A, Lipovova, P, Podzimek, T, Matousek, J, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-06-29 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Plant multifunctional nuclease TBN1 with unexpected phospholipase activity: structural study and reaction-mechanism analysis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
