7WLG
 
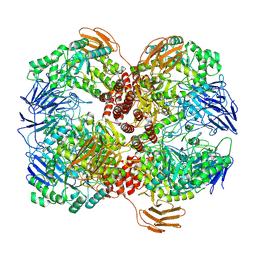 | | Cryo-EM structure of GH31 alpha-1,3-glucosidase from Lactococcus lactis subsp. cremoris | | Descriptor: | Alpha-xylosidase | | Authors: | Ikegaya, M, Moriya, T, Adachi, N, Kawasaki, M, Park, E.Y, Miyazaki, T. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis of the strict specificity of a bacterial GH31 alpha-1,3-glucosidase for nigerooligosaccharides.
J.Biol.Chem., 298, 2022
|
|
7WAF
 
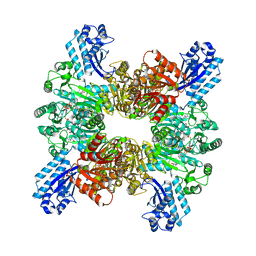 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7WAE
 
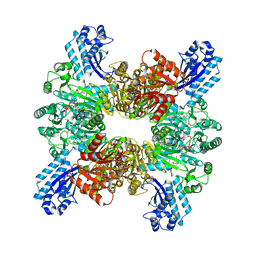 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7EH7
 
 | | Cryo-EM structure of the octameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
7EH8
 
 | | Cryo-EM structure of the hexameric state of C-phycocyanin from Thermoleptolyngbya sp. O-77 | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Minato, T, Teramoto, T, Adachi, N, Hung, N.K, Yamada, K, Kawasaki, M, Akutsu, M, Moriya, T, Senda, T, Ogo, S, Kakuta, Y, Yoon, K.S. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Non-conventional octameric structure of C-phycocyanin.
Commun Biol, 4, 2021
|
|
3VHX
 
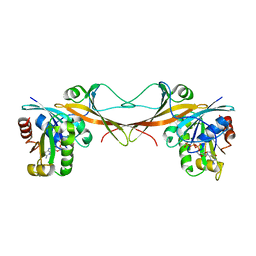 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
7XM1
 
 | | Cryo-EM structure of mTIP60-Ba (metal-ion induced TIP60 (K67E) complex with barium ions | | Descriptor: | BARIUM ION, TIP60 K67E mutant | | Authors: | Ohara, N, Kawakami, N, Arai, R, Adachi, N, Moriya, T, Kawasaki, M, Miyamoto, K. | | Deposit date: | 2022-04-24 | | Release date: | 2023-01-04 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J.Am.Chem.Soc., 145, 2023
|
|
3WXA
 
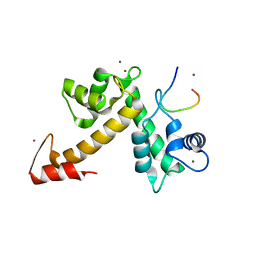 | | X-ray crystal structural analysis of the complex between ALG-2 and Sec31A peptide | | Descriptor: | Programmed cell death protein 6, Protein transport protein Sec31A, ZINC ION | | Authors: | Takahashi, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Int J Mol Sci, 16, 2015
|
|
2ZAM
 
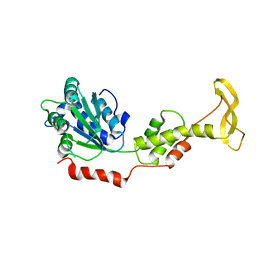 | | Crystal structure of mouse SKD1/VPS4B apo-form | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZAO
 
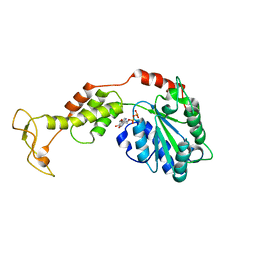 | | Crystal structure of mouse SKD1/VPS4B ADP-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZN9
 
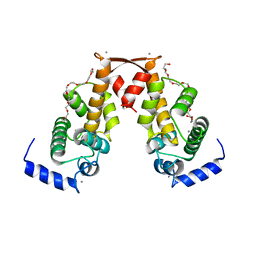 | | Crystal structure of Ca2+-bound form of des3-20ALG-2 | | Descriptor: | CALCIUM ION, DODECAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZNE
 
 | | Crystal structure of Zn2+-bound form of des3-23ALG-2 complexed with Alix ABS peptide | | Descriptor: | 16-meric peptide from Programmed cell death 6-interacting protein, Programmed cell death protein 6, SODIUM ION, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZN8
 
 | | Crystal structure of Zn2+-bound form of ALG-2 | | Descriptor: | Programmed cell death protein 6, SODIUM ION, ZINC ION | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
2ZAN
 
 | | Crystal structure of mouse SKD1/VPS4B ATP-form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
2ZND
 
 | | Crystal structure of Ca2+-free form of des3-20ALG-2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHATE ION, Programmed cell death protein 6, ... | | Authors: | Suzuki, H, Kawasaki, M, Inuzuka, T, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ca(2+)-Dependent Formation of ALG-2/Alix Peptide Complex: Ca(2+)/EF3-Driven Arginine Switch Mechanism
Structure, 16, 2008
|
|
7CN0
 
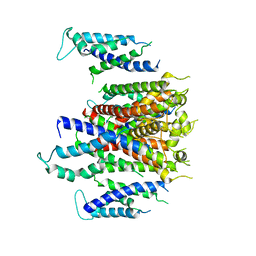 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CN1
 
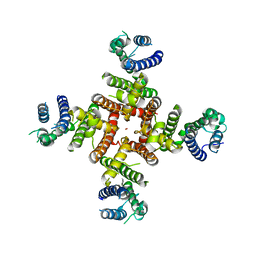 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | Descriptor: | POTASSIUM ION, potassium channel | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
2ZRS
 
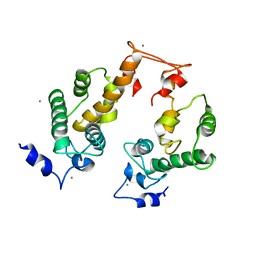 | | Crystal structure of Ca2+-bound form of des3-23ALG-2 | | Descriptor: | CALCIUM ION, Programmed cell death protein 6 | | Authors: | Suzuki, H, Kawasaki, M, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallization and X-ray diffraction analysis of N-terminally truncated human ALG-2
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
2ZVO
 
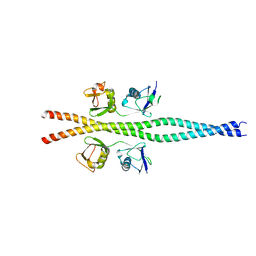 | | NEMO CoZi domain in complex with diubiquitin in C2 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
2ZVN
 
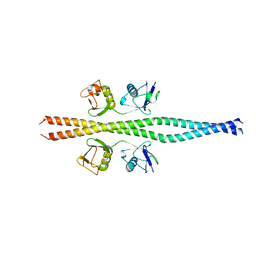 | | NEMO CoZi domain incomplex with diubiquitin in P212121 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
3AI5
 
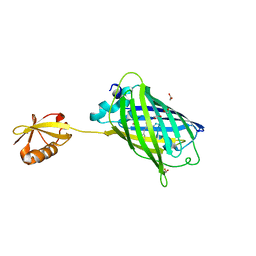 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AAJ
 
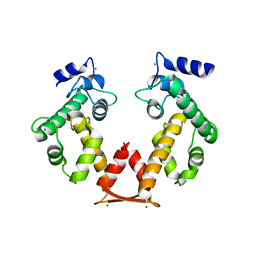 | | Crystal structure of Ca2+-bound form of des3-23ALG-2deltaGF122 | | Descriptor: | CALCIUM ION, Programmed cell death protein 6 | | Authors: | Suzuki, H, Inuzuka, T, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
3AAK
 
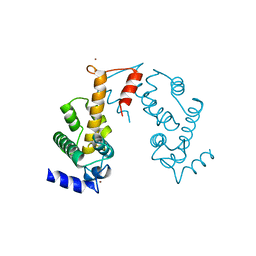 | | Crystal structure of Zn2+-bound form of des3-20ALG-2F122A | | Descriptor: | Programmed cell death protein 6, ZINC ION | | Authors: | Inuzuka, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2ZRT
 
 | | Crystal structure of Zn2+-bound form of des3-23ALG-2 | | Descriptor: | Programmed cell death protein 6, ZINC ION | | Authors: | Suzuki, H, Kawasaki, M, Kakiuchi, T, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystallization and X-ray diffraction analysis of N-terminally truncated human ALG-2
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
