6SQW
 
 | | Mouse dCTPase in complex with 5-Me-dCMP | | Descriptor: | 5-METHYL-2'-DEOXY-CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, dCTP pyrophosphatase 1 | | Authors: | Scaletti, E.R, Claesson, M, Helleday, H, Jemth, A.S, Stenmark, P. | | Deposit date: | 2019-09-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The First Structure of an Active Mammalian dCTPase and its Complexes With Substrate Analogs and Products.
J.Mol.Biol., 432, 2020
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
4C9X
 
 | | Crystal structure of NUDT1 (MTH1) with S-crizotinib | | Descriptor: | 3-[(1S)-1-(2,6-DICHLORO-3-FLUOROPHENYL)ETHOXY]-5-(1-PIPERIDIN-4-YLPYRAZOL-4-YL)PYRIDIN-2-AMINE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
7B67
 
 | | Structure of NUDT15 V18_V19insGV Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7AYY
 
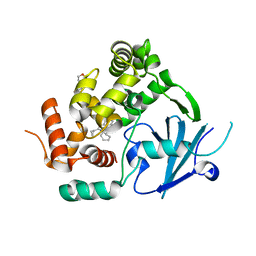 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
7AYZ
 
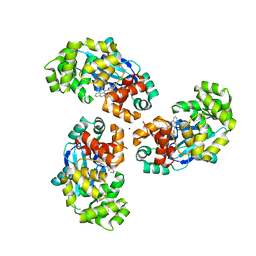 | |
7AZ0
 
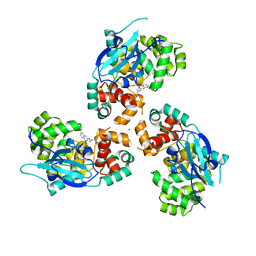 | |
7B65
 
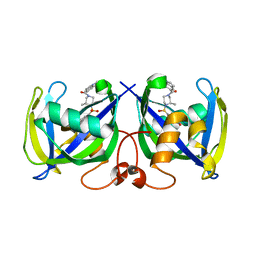 | | Structure of NUDT15 R139C Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B63
 
 | | Structure of NUDT15 in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B66
 
 | | Structure of NUDT15 R139H Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
7B64
 
 | | Structure of NUDT15 V18I Mutant in complex with TH7755 | | Descriptor: | (R)-6-((2-methyl-4-(1-methyl-1H-indole-5-carbonyl)piperazin-1-yl)sulfonyl)benzo[d]oxazol-2(3H)-one, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of NUDT15 variants enabled by a potent inhibitor reveal the structural basis for thiopurine sensitivity.
J.Biol.Chem., 296, 2021
|
|
2RF5
 
 | | Crystal structure of human tankyrase 1- catalytic PARP domain | | Descriptor: | GLYCEROL, Tankyrase-1, ZINC ION | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Zinc binding catalytic domain of human tankyrase 1.
J.Mol.Biol., 379, 2008
|
|
5NQR
 
 | | Potent inhibitors of NUDT5 silence hormone signaling in breast cancer | | Descriptor: | 8-(dimethylamino)-1,3-dimethyl-7-[[5-(3-methylphenyl)-1,3,4-oxadiazol-2-yl]methyl]purine-2,6-dione, ADP-sugar pyrophosphatase | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2017-04-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Nat Commun, 9, 2018
|
|
3CE0
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
3C4H
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor DR2313 | | Descriptor: | 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Poly(ADP-ribose) polymerase 3 | | Authors: | Lehtio, L, Collins, R, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
7QEI
 
 | | Structure of human MTHFD2L in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Probable bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase 2 | | Authors: | Gustafsson, R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of Human MTHFD2L and Its Implications for the Development of Isoform-Selective Inhibitors.
Chemmedchem, 17, 2022
|
|
4XHU
 
 | |
4XHT
 
 | |
4XHW
 
 | |
5LOU
 
 | | human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22, PHOSPHATE ION | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2016-08-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human NUDT22 Is a UDP-Glucose/Galactose Hydrolase Exhibiting a Unique Structural Fold.
Structure, 26, 2018
|
|
5LOR
 
 | | human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2016-08-09 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structural and functional studies of human NUDT22
To Be Published
|
|
5NWH
 
 | | Potent inhibitors of NUDT5 silence hormone signaling in breast cancer | | Descriptor: | 7-[[5-(3,4-dichlorophenyl)-1,3,4-oxadiazol-2-yl]methyl]-1,3-dimethyl-8-piperazin-1-yl-purine-2,6-dione, ADP-sugar pyrophosphatase | | Authors: | Carter, M, Stenmark, P. | | Deposit date: | 2017-05-06 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeted NUDT5 inhibitors block hormone signaling in breast cancer cells.
Nat Commun, 9, 2018
|
|
