7RE3
 
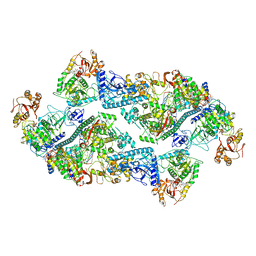 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
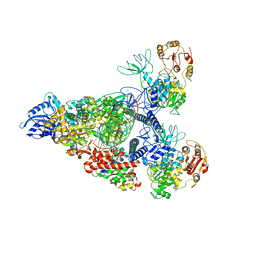 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
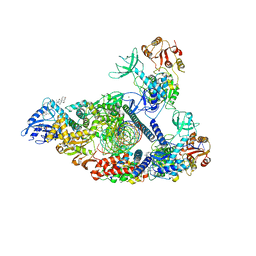 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
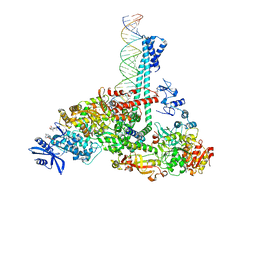 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
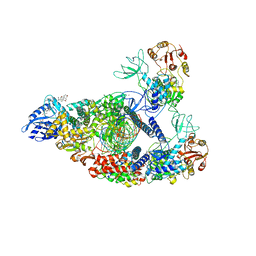 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2Q1Z
 
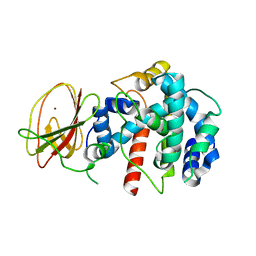 | |
4XLP
 
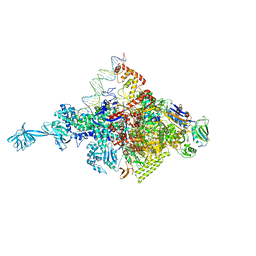 | |
4XSY
 
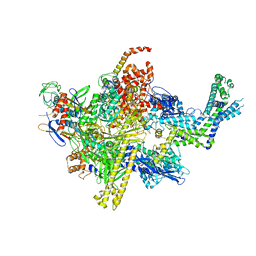 | | Crystal structure of CBR 9379 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 3-{[(2,6-dichlorophenyl)carbamoyl]amino}-N-hydroxy-N'-phenyl-5-(trifluoromethyl)benzenecarboximidamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.007 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3TBI
 
 | |
3UGP
 
 | |
3UGO
 
 | |
1BDF
 
 | |
3NIB
 
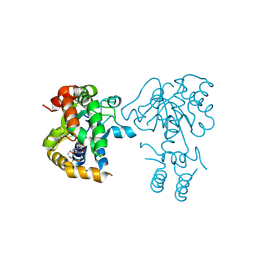 | | Teg14 Apo | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Teg14 | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The 2.7 A resolution structure of the glycopeptide sulfotransferase Teg14
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NRX
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved non-motor microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
2AUJ
 
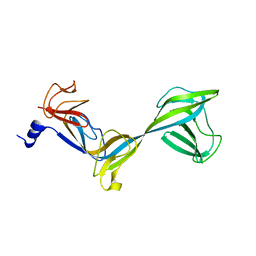 | | Structure of Thermus aquaticus RNA polymerase beta'-subunit insert | | Descriptor: | DNA-directed RNA polymerase beta' chain | | Authors: | Chlenov, M, Masuda, S, Murakami, K.S, Nikiforov, V, Darst, S.A, Mustaev, A. | | Deposit date: | 2005-08-27 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function of Lineage-specific Sequence Insertions in the Bacterial RNA Polymerase beta' Subunit
J.Mol.Biol., 353, 2005
|
|
2AUK
 
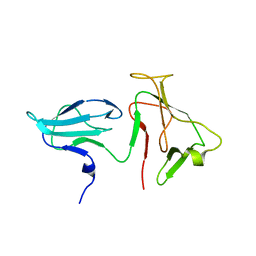 | | Structure of E. coli RNA polymerase beta' G/G' insert | | Descriptor: | DNA-directed RNA polymerase beta' chain | | Authors: | Chlenov, M, Masuda, S, Murakami, K.S, Nikiforov, V, Darst, S.A, Mustaev, A. | | Deposit date: | 2005-08-28 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Lineage-specific Sequence Insertions in the Bacterial RNA Polymerase beta' Subunit
J.Mol.Biol., 353, 2005
|
|
4X8K
 
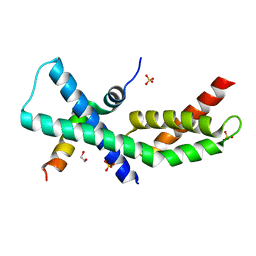 | | Mycobacterium tuberculosis RbpA-SID in complex with SigmaA domain 2 | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor SigA, RNA polymerase-binding protein RbpA, ... | | Authors: | Hubin, E.A, Flack, J.E, Tabib-Salazar, A, Paget, M.S, Darst, S.A, Campbell, E.A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NRY
 
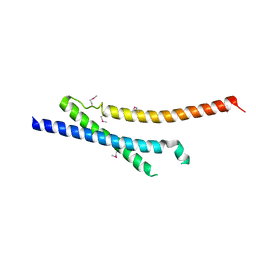 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-07-01 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
4XLS
 
 | |
4XLR
 
 | |
4XLN
 
 | |
4XAX
 
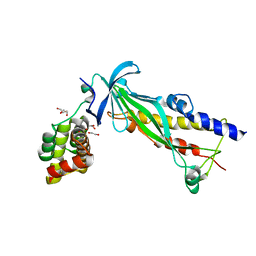 | | Crystal structure of Thermus thermophilus CarD in complex with the Thermus aquaticus RNA polymerase beta1 domain | | Descriptor: | 1,2-ETHANEDIOL, CarD, DNA-directed RNA polymerase subunit beta domain 1 | | Authors: | Chen, J, Bae, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex.
Elife, 4, 2015
|
|
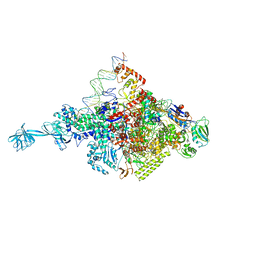
4XLQ
 
 | |
4XSZ
 
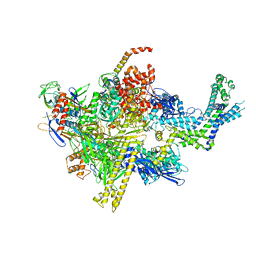 | | Crystal structure of CBR 9393 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 4-[3-(4-fluorophenyl)-1H-pyrazol-4-yl]-N-[2-(piperazin-1-yl)ethyl]-2-(trifluoromethyl)aniline, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.683 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XSX
 
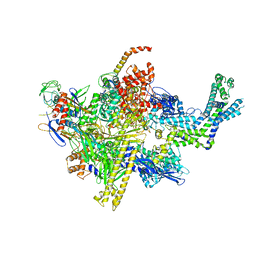 | | Crystal structure of CBR 703 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.708 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
