7LML
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 6-iodanyl-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
5DBR
 
 | |
5DQO
 
 | |
5E7N
 
 | | Crystal Structure of RPA70N in complex with VU0085636 | | Descriptor: | 2-({3-[(4-bromophenyl)sulfamoyl]-4-methylbenzoyl}amino)benzoic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Gilston, B.A, Patrone, J.D, Pelz, N.F, Bates, B.S, Souza-Fagundes, E.M, Vangamudi, B, Camper, D, Kuznetsov, A, Browning, C.F, Feldkamp, M.D, Olejniczak, E.T, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2015-10-12 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Identification and Optimization of Anthranilic Acid Based Inhibitors of Replication Protein A.
Chemmedchem, 11, 2016
|
|
1KCY
 
 | | NMR solution structure of apo calbindin D9k (F36G + P43M mutant) | | Descriptor: | calbindin D9k | | Authors: | Nelson, M.R, Thulin, E, Fagan, P.A, Forsen, S, Chazin, W.J. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The EF-hand domain: a globally cooperative structural unit.
Protein Sci., 11, 2002
|
|
2AMI
 
 | | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | | Descriptor: | Caltractin | | Authors: | Hu, H.T, Fagan, P.A, Bunick, C.G, Sheehan, J.H, Chazin, W.J. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function.
J.Biol.Chem., 281, 2006
|
|
2BAY
 
 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
1D1O
 
 | |
2JTT
 
 | |
2KEB
 
 | | NMR solution structure of the N-terminal domain of the DNA polymerase alpha p68 subunit | | Descriptor: | DNA polymerase subunit alpha B | | Authors: | Huang, H, Weiner, B.E, Zhang, H, Fuller, B.E, Gao, Y, Wile, B.M, Chazin, W.J, Fanning, E. | | Deposit date: | 2009-01-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a DNA polymerase alpha-primase domain that docks on the SV40 helicase and activates the viral primosome.
J.Biol.Chem., 285, 2010
|
|
2KBI
 
 | |
2KWQ
 
 | | Mcm10 C-terminal DNA binding domain | | Descriptor: | Protein MCM10 homolog, ZINC ION | | Authors: | Robertson, P.D, Chagot, B, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif.
J.Biol.Chem., 285, 2010
|
|
4OU0
 
 | |
4O0A
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)-2-chlorophenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejniczak, E.T, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-12-13 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J. Med. Chem., 56, 2013
|
|
4R4Q
 
 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4I
 
 | | Structure of RPA70N in complex with 5-(4-((6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl]methyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4O
 
 | | Crystal structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)benzyl)carbamothioyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]carbamothioyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4T
 
 | | Crystal Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)phenylthioamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)phenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4C
 
 | | Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)-2-chlorobenzamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({[4-(5-carboxyfuran-2-yl)-2-chlorobenzoyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
1CDN
 
 | |
1B1G
 
 | |
1CLB
 
 | |
1A03
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF CA2+-BOUND CALCYCLIN: IMPLICATIONS FOR CA2+-SIGNAL TRANSDUCTION BY S100 PROTEINS, NMR, 20 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, CA2+) | | Authors: | Sastry, M, Ketchem, R.R, Crescenzi, O, Weber, C, Lubienski, M.J, Hidaka, H, Chazin, W.J. | | Deposit date: | 1997-12-08 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins.
Structure, 6, 1998
|
|
4IJH
 
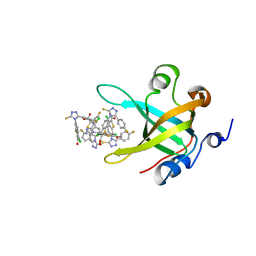 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | 3-chloro-6-[3-(4-fluorophenyl)-5-sulfanyl-4H-1,2,4-triazol-4-yl]-1-benzothiophene-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
4IJL
 
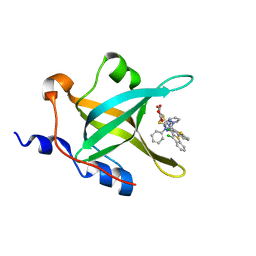 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, {[5-(3-chloro-1-benzothiophen-2-yl)-4-phenyl-4H-1,2,4-triazol-3-yl]sulfanyl}acetic acid | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
