3MHT
 
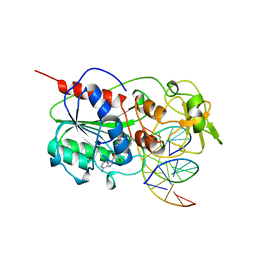 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH UNMODIFIED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | O'Gara, M, Klimasauskas, S, Roberts, R.J, Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
4F7Z
 
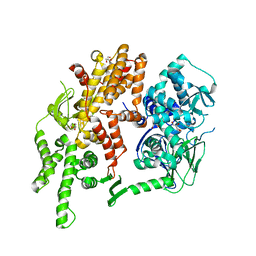 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
3FPD
 
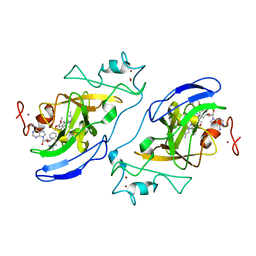 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3SWR
 
 | |
4RNW
 
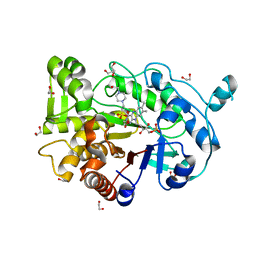 | | Truncated version of the G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNX
 
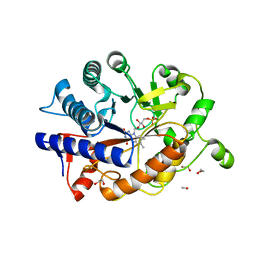 | | K154 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1 | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNU
 
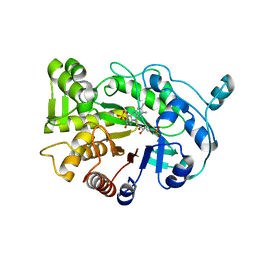 | | G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNV
 
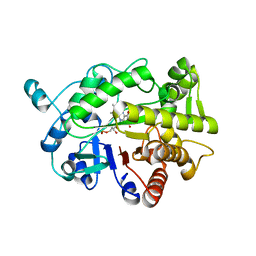 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
3EU9
 
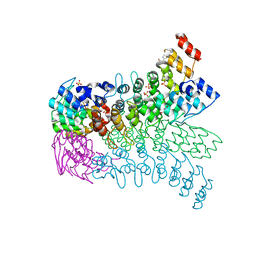 | | The ankyrin repeat domain of Huntingtin interacting protein 14 | | Descriptor: | GLYCEROL, HISTIDINE, Huntingtin-interacting protein 14, ... | | Authors: | Gao, T, Collins, R.E, Horton, J.R, Zhang, R, Zhang, X, Cheng, X. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding.
Proteins, 76, 2009
|
|
4QW2
 
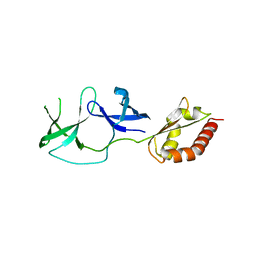 | | FMRP N-terminal domain (R138Q) | | Descriptor: | 1,2-ETHANEDIOL, Fragile X mental retardation protein 1, LEAD (II) ION | | Authors: | Myrick, L.K, Hashimoto, H, Cheng, X, Warren, S.T. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Hum.Mol.Genet., 24, 2015
|
|
4QVZ
 
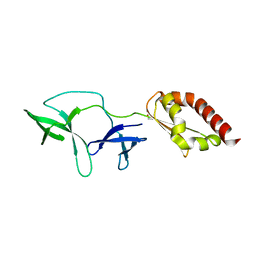 | | FMRP N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Fragile X mental retardation protein 1 | | Authors: | Myrick, L.K, Hashimoto, H, Cheng, X, Warren, S.T. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Hum.Mol.Genet., 24, 2015
|
|
6B0R
 
 | |
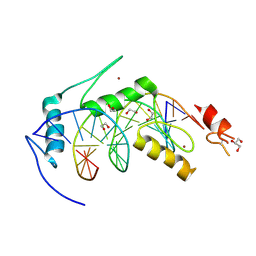
6BLW
 
 | |
6B0Q
 
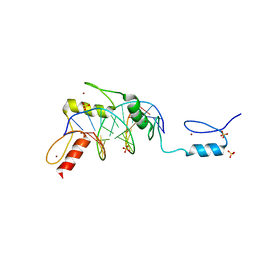 | | Zinc finger Domain of WT1(-KTS form) with 13+1mer Oligonucleotide with 3' Triplet TGT | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*GP*GP*AP*GP*TP*GP*TP*T)-3'), SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-09-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites.
Nucleic Acids Res., 46, 2018
|
|
6B0O
 
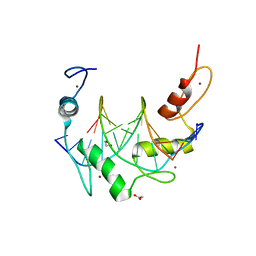 | | Zinc finger Domain of WT1(-KTS form) with 12+1mer Oligonucleotide with 3' Triplet TGT | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*TP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-09-14 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites.
Nucleic Acids Res., 46, 2018
|
|
6B0P
 
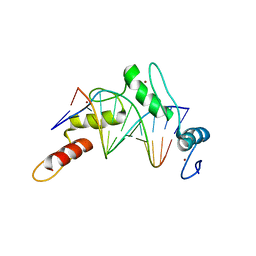 | | Zinc finger Domain of WT1(-KTS form) with 12+1mer Oligonucleotide with 3' Triplet GGT | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*AP*CP*CP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*TP*GP*GP*GP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-09-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Role for first zinc finger of WT1 in DNA sequence specificity: Denys-Drash syndrome-associated WT1 mutant in ZF1 enhances affinity for a subset of WT1 binding sites.
Nucleic Acids Res., 46, 2018
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPC
 
 | | The second RBD of SARS-CoV-2 Omicron Variant in complexed with RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
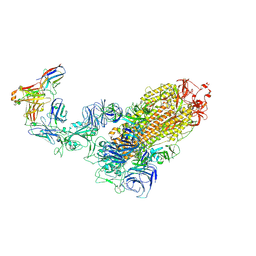 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPF
 
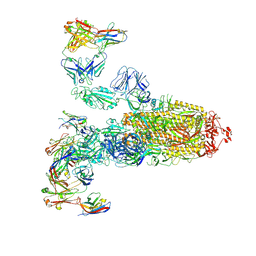 | | SARS-CoV-2 Omicron Variant S Trimer complexed with three JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPE
 
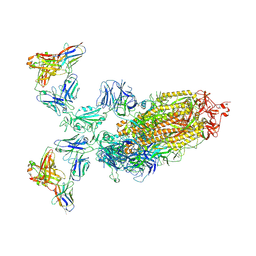 | | SARS-CoV-2 Omicron Variant S Trimer complexed with two JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WRV
 
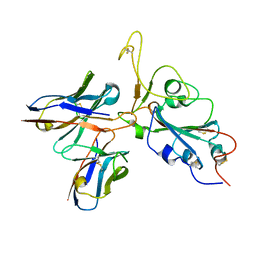 | | The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S | | Descriptor: | JMB2002 Fab heavy chain, JMB2002 Fab light chain, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
3PU8
 
 | | PHF2 Jumonji-NOG-Fe(II) complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
