8UVP
 
 | | Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up) | | Descriptor: | 2-[(4P)-4-(4-{[(4P)-5-(cyclohexylsulfanyl)-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}-2,5-difluorophenyl)-2H-1,2,3-triazol-2-yl]-1-[(2R,6S)-2,6-dimethylmorpholin-4-yl]ethan-1-one, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
8UV2
 
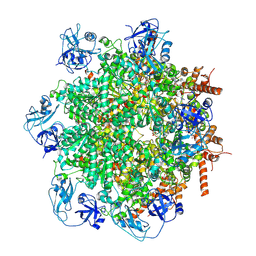 | | Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer) | | Descriptor: | 3-(4-{[(4P)-5-{[(1R)-cyclohex-2-en-1-yl]sulfanyl}-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}-2,6-difluorophenyl)prop-2-yn-1-yl (1-methylpiperidin-4-yl)carbamate, ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2023-11-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
8UVO
 
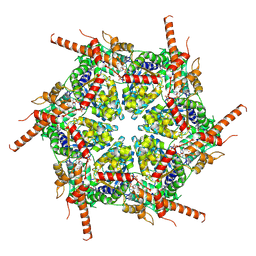 | | Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-[3-(2,5-difluoro-4-{[(4M)-5-(hexylsulfanyl)-4-(pyridin-3-yl)-4H-1,2,4-triazol-3-yl]methoxy}phenyl)prop-2-yn-1-yl]propanamide, Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, DeVore, K, Chiu, P.-L. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Mechanism of allosteric inhibition of human p97/VCP ATPase and its disease mutant by triazole inhibitors.
Commun Chem, 7, 2024
|
|
8V6V
 
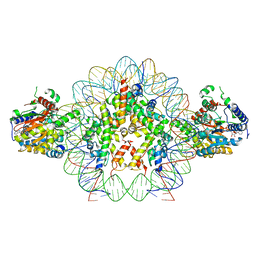 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V4Y
 
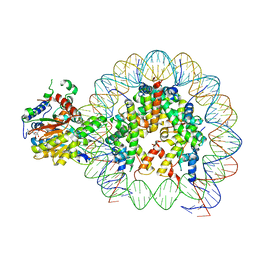 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Histone H2A type 1, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V7L
 
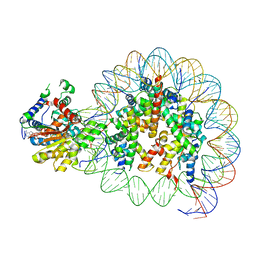 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8Y6U
 
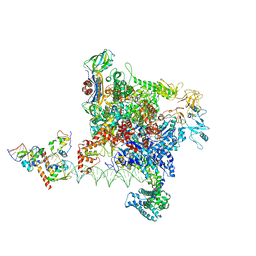 | |
8WSP
 
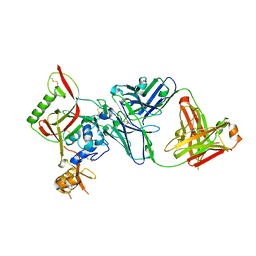 | | Crystal structure of SFTSV Gn and antibody SF5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab5-H, Ab5-L, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | Descriptor: | CalU16 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
8WSU
 
 | | Crystal structure of SFTSV Gc and antibody | | Descriptor: | Ab-H, Ab-L, Glycoprotein C | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSN
 
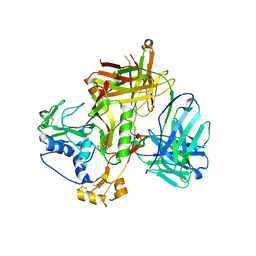 | | Crystal structure of SFTSV Gn and antibody SF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab1-H, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4MMG
 
 | | crystal structure of YafQ mutant H87Q from E.coli | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4MMJ
 
 | | crystal structure of YafQ from E.coli strain BL21(DE3) | | Descriptor: | Addiction module toxin, RelE/StbE family, SULFATE ION | | Authors: | Liang, Y, Gao, Z, Liu, Q, Dong, Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4ML2
 
 | | Crystal structure of wild-type YafQ | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
2NQ8
 
 | | Malarial enoyl acyl ACP reductase bound with INH-NAD adduct | | Descriptor: | Enoyl-acyl carrier reductase, ISONICOTINIC-ACETYL-NICOTINAMIDE-ADENINE DINUCLEOTIDE | | Authors: | Freundlich, J.S, Yu, M, Lucumi, E, Kuo, M, Tsai, H.C, Valderramos, J.C, Karagyozov, L, Jacobs Jr, W.R, Schiehser, G.A, Fidock, D.A, Jacobus, D.P, Sacchettini, J.C. | | Deposit date: | 2006-10-30 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
4ML0
 
 | | Crystal structure of E.coli DinJ-YafQ complex | | Descriptor: | Predicted antitoxin of YafQ-DinJ toxin-antitoxin system, Predicted toxin of the YafQ-DinJ toxin-antitoxin system, SULFATE ION | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-06 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H48
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
7D99
 
 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D90
 
 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
2OL4
 
 | | Crystal structure of plasmodium falciparum enoyl ACP reductase with triclosan reductase | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(3-PHENYLPROPYL)PHENOL, Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tsai, H. | | Deposit date: | 2007-01-18 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
2OP0
 
 | | Crystal structure of plasmodium falciparum enoyl ACP reductase with triclosan reductase | | Descriptor: | 2-[4-(AMINOMETHYL)-2-CHLOROPHENOXY]-5-PYRIDIN-2-YLPHENOL, Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tsai, H. | | Deposit date: | 2007-01-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
2OOS
 
 | | Crystal structure of plasmodium falciparum enoyl ACP reductase with triclosan reductase | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(2-PHENYLETHYL)PHENOL, Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tsai, H. | | Deposit date: | 2007-01-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structural analysis of Plasmodium falciparum enoyl acyl carrier protein reductase as a pathway toward the optimization of triclosan antimalarial efficacy
J.Biol.Chem., 282, 2007
|
|
