8X5D
 
 | |
6AKW
 
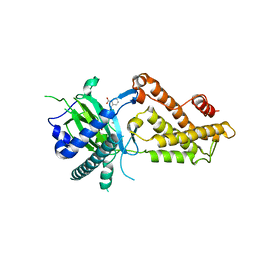 | | Crystal structure of RNA dioxygenase bound with an inhibitor | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2018-09-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia.
Cancer Cell, 35, 2019
|
|
3NYC
 
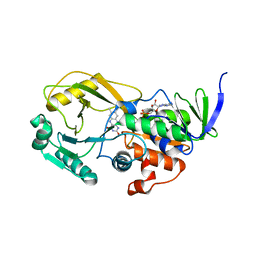 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase | | Descriptor: | (2E)-5-[(diaminomethylidene)amino]-2-iminopentanoic acid, D-Arginine Dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fu, G, Weber, I.T. | | Deposit date: | 2010-07-14 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Conformational changes and substrate recognition in Pseudomonas aeruginosa D-arginine dehydrogenase.
Biochemistry, 49, 2010
|
|
3NYF
 
 | |
7LBC
 
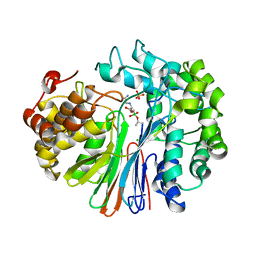 | | Structure of human GGT1 in complex with Lnt2-65 compound | | Descriptor: | (2R)-2-[(2-aminoethyl)amino]-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-07 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
6AZR
 
 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
7LD9
 
 | | Structure of human GGT1 in complex with ABBA | | Descriptor: | (2R)-2-amino-4-boronobutanoic acid, (2S)-2-amino-4-boronobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Terzyan, S.S, Hanigan, M. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design and evaluation of novel analogs of 2-amino-4-boronobutanoic acid (ABBA) as inhibitors of human gamma-glutamyl transpeptidase.
Bioorg.Med.Chem., 73, 2022
|
|
3NYE
 
 | |
6CCK
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3-(3-chlorophenyl)-3-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)propanenitrile | | Descriptor: | (3R)-3-(3-chlorophenyl)-3-[(5-methyl-7-oxo-6,7-dihydro[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]propanenitrile, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCN
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-2,4-dihydroxy-N-(2-(4-hydroxy-1H-benzo[d]imidazol-2-yl)ethyl)-3,3-dimethylbutanamide | | Descriptor: | (2R)-2,4-dihydroxy-N-[2-(7-hydroxy-1H-benzimidazol-2-yl)ethyl]-3,3-dimethylbutanamide, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6D2R
 
 | | HLA-B*57:01 presenting GSFDYSGVHLW | | Descriptor: | Beta-2-microglobulin, GLY-SER-PHE-ASP-TYR-SER-GLY-VAL-HIS-LEU-TRP, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
6D2B
 
 | | HLA-B*57:01 presenting LSDSTARDVTW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
6CCL
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 1-benzyl-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1-benzyl-1H-imidazo[4,5-b]pyridine, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCM
 
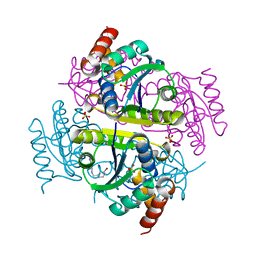 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCS
 
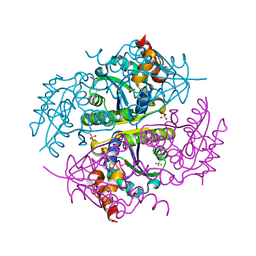 | |
7LM2
 
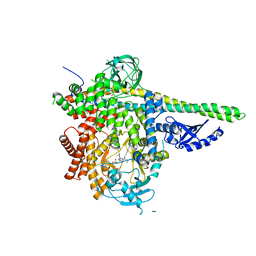 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 3C | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ... | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Projected Dose Optimization of Amino- and Hydroxypyrrolidine Purine PI3K delta Immunomodulators.
J.Med.Chem., 64, 2021
|
|
6CCO
 
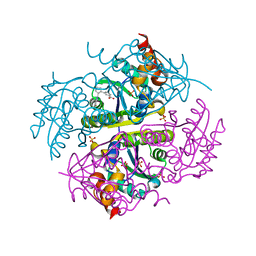 | |
6CCQ
 
 | |
4PV5
 
 | | Crystal structure of mouse glyoxalase I in complexed with 18-beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhang, H, Zhai, J, Zhang, L.P, Zhao, Y.N, Li, C, Hu, X.P. | | Deposit date: | 2014-03-15 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for 18-beta-glycyrrhetinic acid as a novel non-GSH analog glyoxalase I inhibitor
Acta Pharmacol.Sin., 36, 2015
|
|
2G1Q
 
 | | crystal structure of KSP in complex with inhibitor 9h | | Descriptor: | (5S)-5-(3-AMINOPROPYL)-3-(2,5-DIFLUOROPHENYL)-N-ETHYL-5-PHENYL-4,5-DIHYDRO-1H-PYRAZOLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-02-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 4: Structure-based design of 5-alkylamino-3,5-diaryl-4,5-dihydropyrazoles as potent, water-soluble inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6D2T
 
 | | HLA-B*57:01 presenting LALLTGVRW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
6D29
 
 | | HLA-B*57:01 presenting TSMSFVPRPW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands.
Sci Immunol, 3, 2018
|
|
2H95
 
 | |
3EYW
 
 | | Crystal structure of the C-terminal domain of E. coli KefC in complex with KefF | | Descriptor: | C-terminal domain of Glutathione-regulated potassium-efflux system protein kefC fused to full length Glutathione-regulated potassium-efflux system ancillary protein kefF, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Roosild, T.P. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | KTN (RCK) Domains Regulate K(+) Channels and Transporters by Controlling the Dimer-Hinge Conformation.
Structure, 17, 2009
|
|
3HPT
 
 | | Crystal structure of human FxA in complex with (S)-2-cyano-1-(2-methylbenzofuran-5-yl)-3-(2-oxo-1-(2-oxo-2-(pyrrolidin-1-yl)ethyl)azepan-3-yl)guanidine | | Descriptor: | 1-cyano-2-(2-methyl-1-benzofuran-5-yl)-3-[(3S)-2-oxo-1-(2-oxo-2-pyrrolidin-1-ylethyl)azepan-3-yl]guanidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Klei, H.E, Ghosh, K, Rushith, A, Kish, K. | | Deposit date: | 2009-06-04 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyanoguanidine-based lactam derivatives as a novel class of orally bioavailable factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
