3KXK
 
 | |
1VFQ
 
 | |
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8E6Q
 
 | | Human TRPM2 ion channel in 1 mM ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6R
 
 | | Human TRPM2 ion channel in 1 mM dADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6S
 
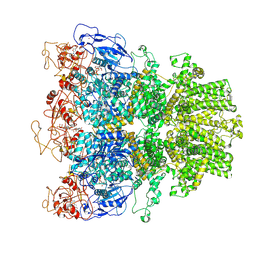 | | Human TRPM2 ion channel in 1 mM dADPR and Ca2+ | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, ... | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6T
 
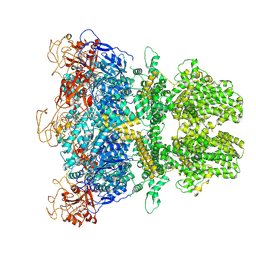 | | Human TRPM2 ion channel in 1 mM BR-ADPR | | Descriptor: | (3R,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4,5-trihydroxypentyl [(2S,3S,4S,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6U
 
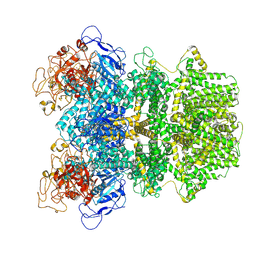 | | Human TRPM2 ion channel in 1 mM F-dADPR | | Descriptor: | Transient receptor potential cation channel subfamily M member 2, [(2R,3S,5R)-5-(6-amino-8-phenyl-9H-purin-9-yl)-3-hydroxyoxolan-2-yl]methyl [(2R,3S,4S,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6V
 
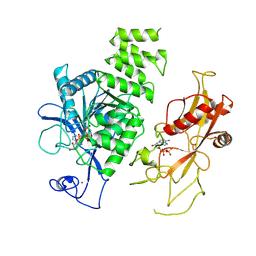 | | MHR1/2 and NUDT9H of human TRPM2 in 1 mM dADPR (local refinement) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2, ZINC ION | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8TAG
 
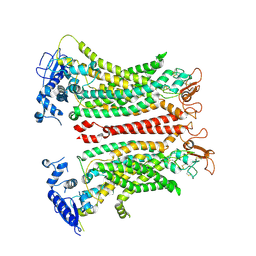 | | TMEM16F, with Calcium and PIP2, no inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Cheng, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TB1
 
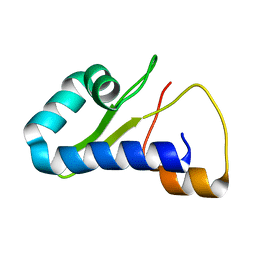 | |
8UF5
 
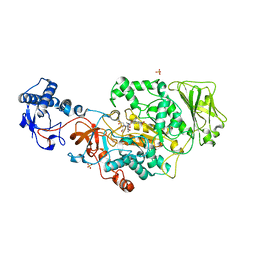 | | Catalytic domain of GtfB in complex with inhibitor G43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2023-10-03 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Discovery of Small Molecule Inhibitors of Cariogenic Virulence.
Sci Rep, 7, 2017
|
|
7F3B
 
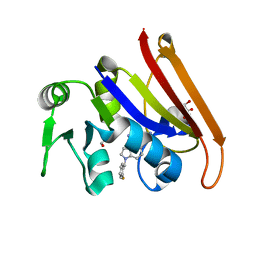 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
4YPG
 
 | |
8URV
 
 | |
3BV2
 
 | | Morpholino pyrrolotriazine P38 Alpha map kinase inhibitor compound 30 | | Descriptor: | 5-methyl-4-[(2-methyl-5-{[(2-morpholin-4-ylpyridin-4-yl)carbonyl]amino}phenyl)amino]-N-(1-phenylethenyl)pyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BV3
 
 | | Morpholino pyrrolotriazine P38 Alpha Map Kinase inhibitor compound 2 | | Descriptor: | 5-methyl-4-[(2-methyl-5-{[(3-morpholin-4-ylphenyl)carbonyl]amino}phenyl)amino]-N-[(1S)-1-phenylethyl]pyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2008-01-04 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Synthesis and SAR of new pyrrolo[2,1-f][1,2,4]triazines as potent p38 alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4ZN2
 
 | | Glycosyl hydrolase from Pseudomonas aeruginosa | | Descriptor: | PslG | | Authors: | Su, T, Liu, S, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix
Cell Res., 25, 2015
|
|
4ZNE
 
 | | IgG1 Fc-FcgammaRI ecd complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Oganesyan, V.Y, Dall'Acqua, W.F. | | Deposit date: | 2015-05-04 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into the interaction of human IgG1 with Fc gamma RI: no direct role of glycans in binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7E80
 
 | | Cryo-EM structure of the flagellar rod with hook and export apparatus from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E81
 
 | | Cryo-EM structure of the flagellar MS ring with FlgB-Dc loop and FliE-helix 1 from Salmonella | | Descriptor: | Flagellar M-ring protein, FlgB-Dc loop, FliE helix 1 | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7E82
 
 | | Cryo-EM structure of the flagellar rod with partial hook from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2021-02-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
