6LBP
 
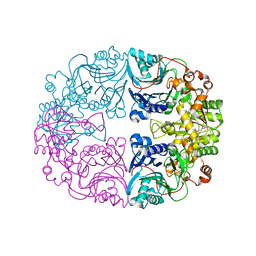 | | Structure of the Glutamine Phosphoribosylpyrophosphate Amidotransferase from Arabidopsis thaliana | | Descriptor: | Amidophosphoribosyltransferase 2, chloroplastic, IRON/SULFUR CLUSTER | | Authors: | Yi, Z, Cao, X, Han, F, Feng, Y. | | Deposit date: | 2019-11-14 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.065 Å) | | Cite: | Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 FromArabidopsis thaliana.
Front Plant Sci, 11, 2020
|
|
6KW4
 
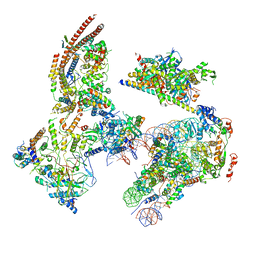 | | The ClassB RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.55 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
4WA7
 
 | | Crystal Structure of a GDP-bound Q61L Oncogenic Mutant of Human GT- Pase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-08-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
5WIH
 
 | |
7CBY
 
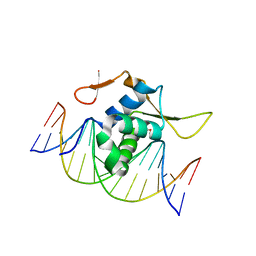 | | Structure of FOXG1 DNA binding domain bound to DBE2 DNA site | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Dai, S.Y, Li, J, Chen, Y.H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural Basis for DNA Recognition by FOXG1 and the Characterization of Disease-causing FOXG1 Mutations.
J.Mol.Biol., 432, 2020
|
|
8HJ4
 
 | |
8WSN
 
 | | Crystal structure of SFTSV Gn and antibody SF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab1-H, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSP
 
 | | Crystal structure of SFTSV Gn and antibody SF5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab5-H, Ab5-L, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSU
 
 | | Crystal structure of SFTSV Gc and antibody | | Descriptor: | Ab-H, Ab-L, Glycoprotein C | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XNG
 
 | |
8XS4
 
 | |
8XW1
 
 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW2
 
 | |
8XW3
 
 | |
8XS5
 
 | |
8XW0
 
 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVY
 
 | |
8X43
 
 | | human KCNQ2-CaM-Ebio1-S1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(4-azanyl-1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-11-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
8XVZ
 
 | |
8XW4
 
 | | Cryo-EM structure of TMEM63B-Digitonin state | | Descriptor: | CSC1-like protein 2 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XS0
 
 | |
8XVX
 
 | |
8XRY
 
 | |
