1MVX
 
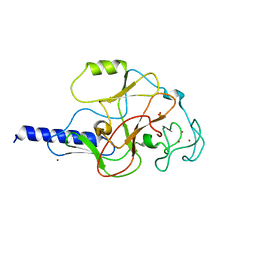 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | CRYPTIC LOCI REGULATOR 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-26 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
1MVH
 
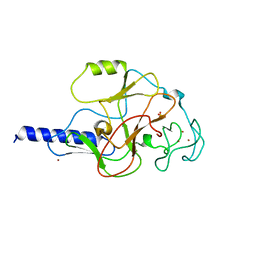 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
1NW3
 
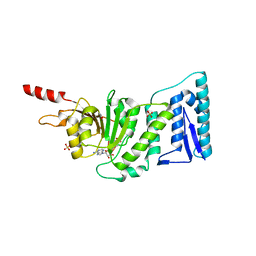 | | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Min, J.R, Feng, Q, Li, Z.H, Zhang, Y, Xu, R.M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase
Cell(Cambridge,Mass.), 112, 2003
|
|
2PQW
 
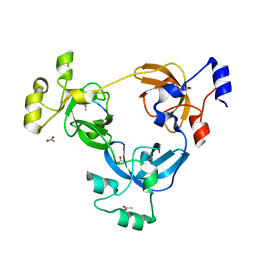 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), trigonal form | | Descriptor: | ACETATE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJD
 
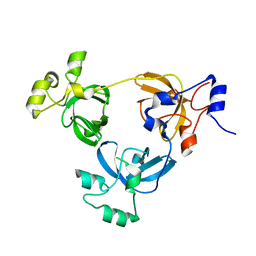 | | Crystal structure of L3MBTL1 protein | | Descriptor: | Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJC
 
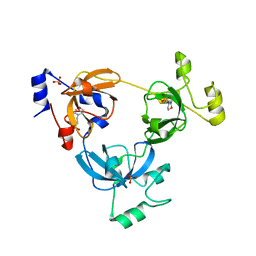 | | Crystal structure of L3MBTL1 protein in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJF
 
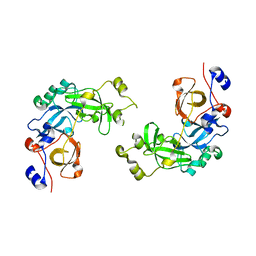 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | Descriptor: | Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RJE
 
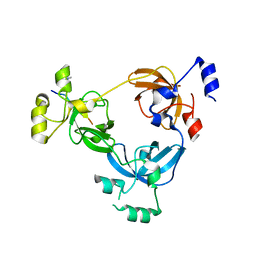 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 17-25), orthorhombic form II | | Descriptor: | CHLORIDE ION, Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6CFI
 
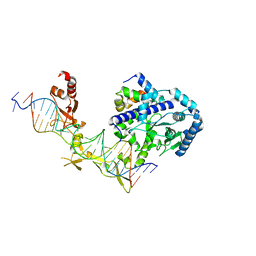 | | Crystal structure of Rad4-Rad23 bound to a 6-4 photoproduct UV lesion | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*(T64)P*TP*GP*GP*AP*TP*GP*TP*TP*GP*AP*GP*TP*CP*A)-3'), DNA repair protein RAD4, DNA('-D(*TP*TP*GP*AP*CP*TP*CP*AP*AP*CP*AP*TP*CP*CP*AP*AP*AP*GP*CP*TP*AP*CP*AP*A)-'), ... | | Authors: | Min, J, Jeffrey, P.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.36241913 Å) | | Cite: | Structure and mechanism of pyrimidine-pyrimidone (6-4) photoproduct recognition by the Rad4/XPC nucleotide excision repair complex.
Nucleic Acids Res., 47, 2019
|
|
2EX4
 
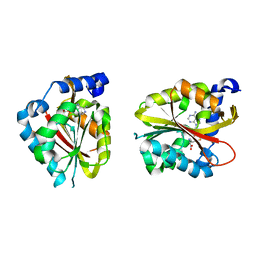 | | Crystal Structure of Human methyltransferase AD-003 in complex with S-adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, adrenal gland protein AD-003 | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-07 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of Human AD-003 protein in complex with S-adenosyl-L-homocysteine
To be Published
|
|
5JOE
 
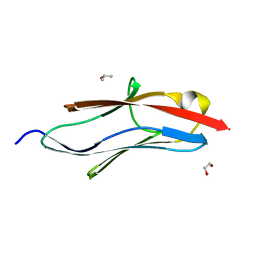 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
5N2P
 
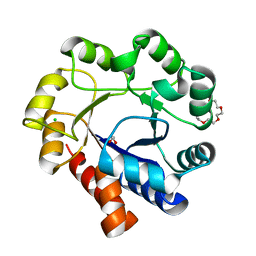 | | Sulfolobus solfataricus Tryptophan Synthase A | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Tryptophan synthase alpha chain | | Authors: | Fleming, J, Mayans, O. | | Deposit date: | 2017-02-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Evolutionary Morphing of Tryptophan Synthase: Functional Mechanisms for the Enzymatic Channeling of Indole.
J.Mol.Biol., 430, 2018
|
|
6HTE
 
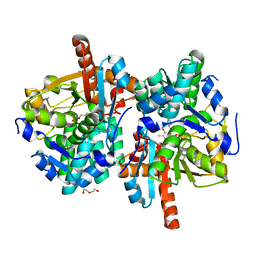 | | Sulfolobus solfataricus Tryptophan Synthase B2a | | Descriptor: | DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase beta chain 2 | | Authors: | Fleming, J, Mayans, O, Bucher, R. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Evolutionary Morphing of Tryptophan Synthase: Functional Mechanisms for the Enzymatic Channeling of Indole.
J.Mol.Biol., 430, 2018
|
|
3KUF
 
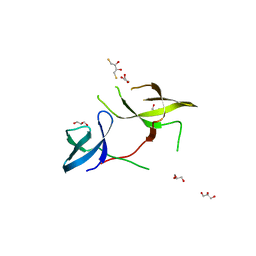 | | The Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Fragile X mental retardation syndrome-related protein 1, GLYCEROL | | Authors: | Bian, C, Guo, Y.H, Adams-Cioaba, M.A, Mackenzie, F, Kozieradzki, I, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2010-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Tudor domains from Fragile X mental retardation syndrome-related protein 1
To be Published
|
|
8J0K
 
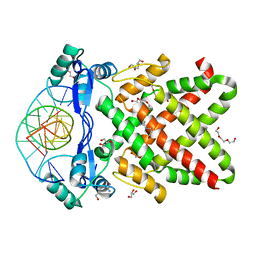 | | Crystal structure of human TFAP2A in complex with DNA | | Descriptor: | DNA (5'-D(*CP*TP*GP*CP*CP*TP*CP*GP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*CP*GP*AP*GP*GP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0Q
 
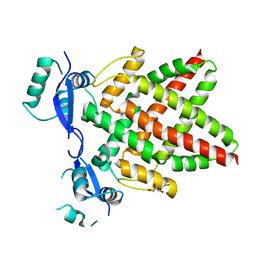 | | Structure of DNA binding domain of human TFAP2B | | Descriptor: | GLYCEROL, Transcription factor AP-2-beta | | Authors: | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0L
 
 | | Structure of DNA binding Domain of Human TFAP2A | | Descriptor: | GLYCEROL, Transcription factor AP-2-alpha | | Authors: | Liu, K, Xiao, Y.Q, Gan, L.Y, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
8J0R
 
 | | Structure of human TFAP2A in complex with DNA | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*CP*CP*TP*CP*AP*GP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*TP*GP*AP*GP*GP*CP*AP*G)-3'), ... | | Authors: | Liu, K, Xiao, Y.Q, Li, W.F, Min, J.R. | | Deposit date: | 2023-04-11 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for specific DNA sequence motif recognition by the TFAP2 transcription factors.
Nucleic Acids Res., 51, 2023
|
|
4LU1
 
 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli, mutant D69A | | Descriptor: | Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
8DIP
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001023030 | | Descriptor: | (2P)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-2-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6V6X
 
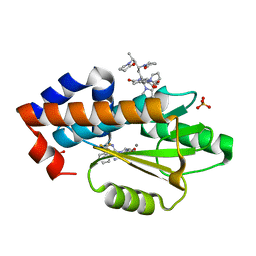 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000988632 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8J85
 
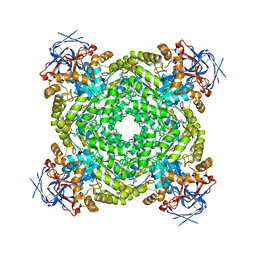 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8JI5
 
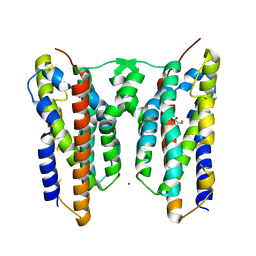 | | Crystal structure of AetD in complex with 5-bromo-L-tryptophan and two Fe2+ | | Descriptor: | 5-bromo-L-tryptophan, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI4
 
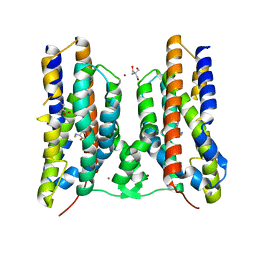 | | Crystal structure of AetD in complex with 5-bromo-L-tryptophan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-bromo-L-tryptophan, AetD, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI3
 
 | | Crystal structure of AetD in complex with 5,7-dibromo-L-tryptophan and two Fe2+ | | Descriptor: | (2S)-2-azanyl-3-[5,7-bis(bromanyl)-1H-indol-3-yl]propanoic acid, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
