5DCQ
 
 | | Crystal structure of bacterial adhesin, FNE from Streptococcus equi spp. equi. | | Descriptor: | FORMIC ACID, Fibronectin-binding protein, artificial repeat proteins (alphaREP3) | | Authors: | Tiouajni, M, Graille, M, van Tilbeurgh, H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural and functional analysis of the fibronectin-binding protein FNE from Streptococcus equi spp. equi.
FEBS J., 281, 2014
|
|
2JG6
 
 | | CRYSTAL STRUCTURE OF A 3-METHYLADENINE DNA GLYCOSYLASE I FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | DNA-3-METHYLADENINE GLYCOSIDASE, ZINC ION | | Authors: | Yan, X, Carter, L.G, Liu, H, Dorward, M, McMahon, S.A, Johnson, K.A, Oke, M, Coote, P.J, Naismith, J.H. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2XNV
 
 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | 2-(2-(4-PHENYLPIPERIDIN-1-YL)ETHYL)-1H-INDOLE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
2XNU
 
 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | 2-(2-(4-PHENYLPIPERIDIN-1-YL)ETHYL)-1H-INDOLE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
7ZL3
 
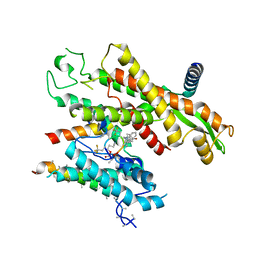 | |
3U6K
 
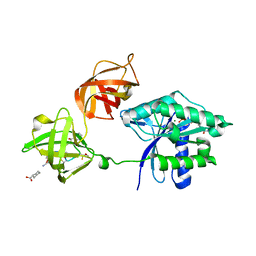 | | Ef-tu (escherichia coli) in complex with nvp-ldk733 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
6TA5
 
 | | OprM-MexA complex from the MexAB-OprM Pseudomonas aeruginosa whole assembly reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
6T7S
 
 | | MexB structure solved by cryo-EM in nanodisc in absence of its protein partners | | Descriptor: | Efflux pump membrane transporter | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
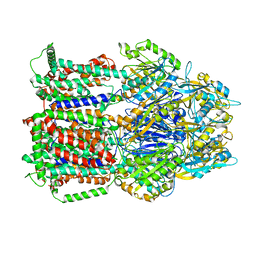
6TA6
 
 | | MexAB assembly of the Pseudomonas MexAB-OprM efflux pump reconstituted in nanodiscs | | Descriptor: | Efflux pump membrane transporter, MexA family multidrug efflux RND transporter periplasmic adaptor subunit, Outer membrane protein OprM | | Authors: | Glavier, M, Schoehn, G, Taveau, J.C, Phan, G, Daury, L, Lambert, O, Broutin, I. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Nat Commun, 11, 2020
|
|
8LPR
 
 | |
6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
2H36
 
 | |
3U6B
 
 | | Ef-tu (escherichia coli) in complex with nvp-ldi028 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
2XQB
 
 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
3UR0
 
 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
7Y3L
 
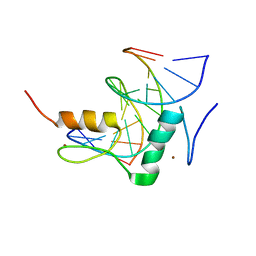 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | Descriptor: | DNA (12-mer), Sal-like protein 3, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3K
 
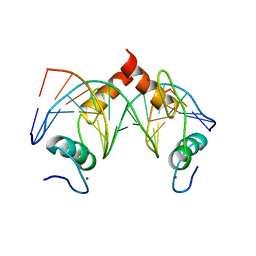 | | Structure of SALL4 ZFC4 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3I
 
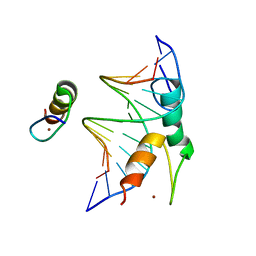 | | Structure of DNA bound SALL4 | | Descriptor: | DNA (12-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
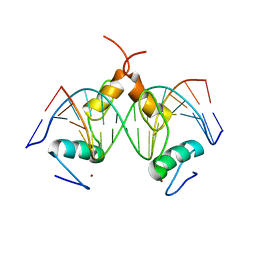 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
3UPF
 
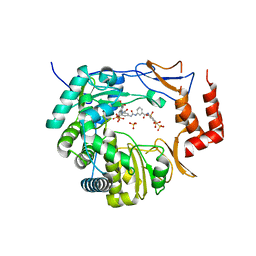 | | Crystal structure of murine norovirus RNA-dependent RNA polymerase bound to NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
2N99
 
 | | Solution structure of the SLURP-2, a secreted isoform of Lynx1 | | Descriptor: | Ly-6/neurotoxin-like protein 1 | | Authors: | Paramonov, A.S, Shenkarev, Z.O, Lyukmanova, E.N, Arseniev, A.S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors.
Sci Rep, 6, 2016
|
|
2NV5
 
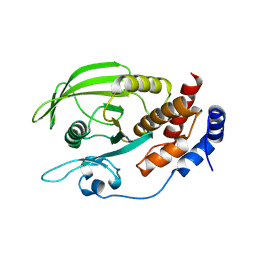 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | protein-tyrosine-phosphatase | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3PQS
 
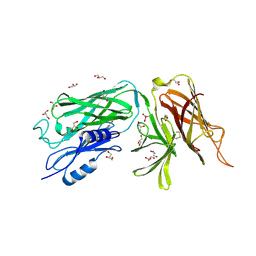 | |
3U2Q
 
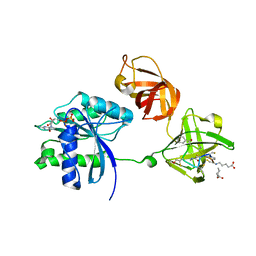 | | EF-Tu (Escherichia coli) in complex with NVP-LFF571 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of LFF571: an investigational agent for Clostridium difficile infection.
J.Med.Chem., 55, 2012
|
|
