8HXB
 
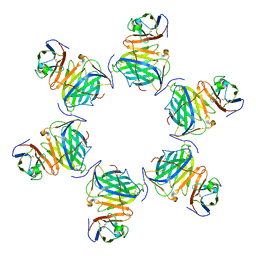 | | Cryo-EM structure of MPXV M2 hexamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXC
 
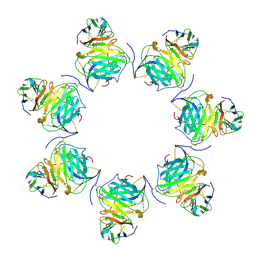 | | Cryo-EM structure of MPXV M2 heptamer in complex with human B7.2 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD86 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
8HXA
 
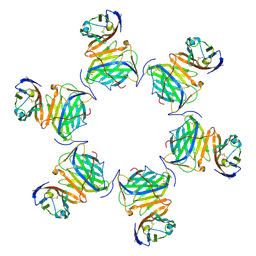 | | Cryo-EM structure of MPXV M2 in complex with human B7.1 | | Descriptor: | NFkB inhibitor, T-lymphocyte activation antigen CD80 | | Authors: | Wang, Y, Yang, S, Zhao, H, Deng, Z. | | Deposit date: | 2023-01-04 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Nat Commun, 14, 2023
|
|
7C7C
 
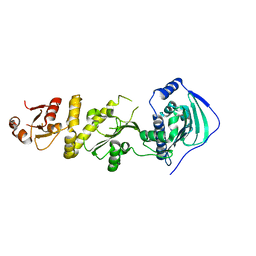 | | Crystal structure of human TRAP1 with SJT104 | | Descriptor: | 2-azanyl-9-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
7C7B
 
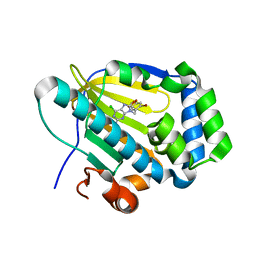 | | Crystal structure of human TRAP1 with SJT009 | | Descriptor: | 2-azanyl-9-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
2GYT
 
 | |
6KWX
 
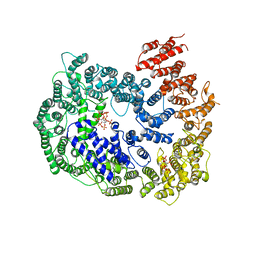 | | cryo-EM structure of human PA200 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, [(1~{S},2~{R},3~{R},4~{S},5~{S},6~{R})-2-[oxidanyl(phosphonooxy)phosphoryl]oxy-3,4,5,6-tetraphosphonooxy-cyclohexyl] phosphono hydrogen phosphate | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
6M39
 
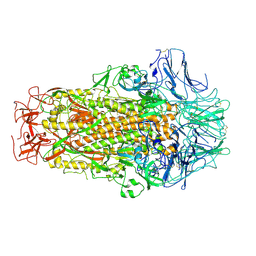 | | Cryo-EM structure of SADS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.
J.Virol., 94, 2020
|
|
4WYH
 
 | |
3VCF
 
 | | SSV1 integrase C-terminal catalytic domain (174-335aa) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-01-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DKS
 
 | | A spindle-shaped virus protein (chymotrypsin treated) | | Descriptor: | Probable integrase | | Authors: | Ouyang, S, Liang, W, Huang, L, Liu, Z.-J. | | Deposit date: | 2012-02-04 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of the C-terminal catalytic domain of SSV1 integrase.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5FAD
 
 | | SAH complex with aKMT from the hyperthermophilic archaeon Sulfolobus islandicus | | Descriptor: | MAGNESIUM ION, Ribosomal protein L11 methyltransferase, putative, ... | | Authors: | Ouyang, S. | | Deposit date: | 2015-12-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | aKMT Catalyzes Extensive Protein Lysine Methylation in the Hyperthermophilic Archaeon Sulfolobus islandicus but is Dispensable for the Growth of the Organism
Mol.Cell Proteomics, 15, 2016
|
|
5FA8
 
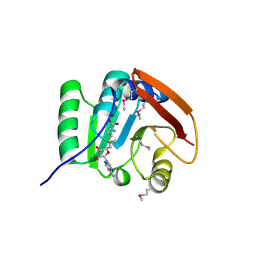 | | SAM complex with aKMT from the hyperthermophilic archaeon Sulfolobus islandicu | | Descriptor: | MAGNESIUM ION, Ribosomal protein L11 methyltransferase, putative, ... | | Authors: | Ouyang, S. | | Deposit date: | 2015-12-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | aKMT Catalyzes Extensive Protein Lysine Methylation in the Hyperthermophilic Archaeon Sulfolobus islandicus but is Dispensable for the Growth of the Organism
Mol.Cell Proteomics, 15, 2016
|
|
6K1T
 
 | |
7W3S
 
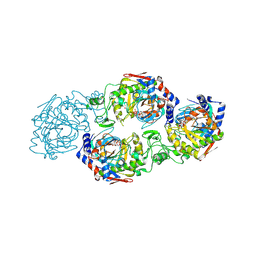 | | The complex structure of Larg1-ADPr from Legionella pneumophila | | Descriptor: | Type IV secretion protein Dot, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Guan, H, Li, P. | | Deposit date: | 2021-11-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Legionella pneumophila temporally regulates the activity of ADP/ATP translocases by reversible ADP-ribosylation.
mLife, 2022
|
|
6KUE
 
 | |
6K11
 
 | |
6KL4
 
 | | Crystal structure of MavC-UBE2N-Ub | | Descriptor: | MavC, Ub, Ubiquitin-conjugating enzyme E2 N | | Authors: | Ouyang, S, Guan, H. | | Deposit date: | 2019-07-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Basis of Ubiquitination Catalyzed by the Bacterial Transglutaminase MavC.
Adv Sci, 7, 2020
|
|
6KWY
 
 | | human PA200-20S complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Proteasome activator complex subunit 4, Proteasome subunit alpha type-1, ... | | Authors: | Ouyang, S, Hongxin, G. | | Deposit date: | 2019-09-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures.
Plos Biol., 18, 2020
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
4OLS
 
 | | The amidase-2 domain of LysGH15 | | Descriptor: | Endolysin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
4OLK
 
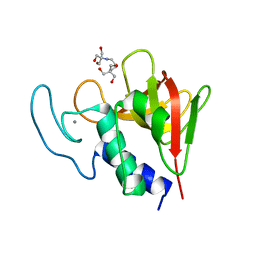 | | The CHAP domain of LysGH15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endolysin | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
5D39
 
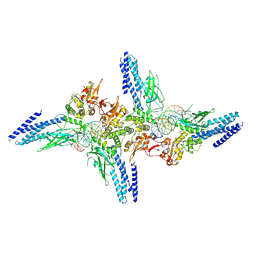 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
