4Z07
 
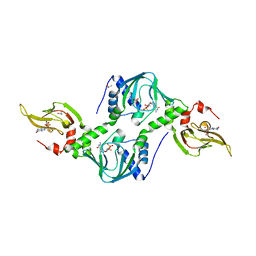 | | Co-crystal structure of the tandem CNB (CNB-A/B) domains of human PKG I beta with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kim, J.J, Reger, A.S, Arold, S.T, Kim, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PKG I:cGMP Complex Reveals a cGMP-Mediated Dimeric Interface that Facilitates cGMP-Induced Activation.
Structure, 24, 2016
|
|
6V6Q
 
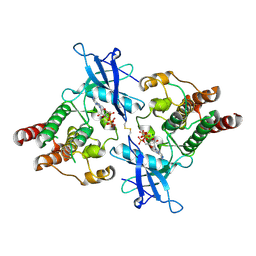 | | Crystal Structure of Monophosphorylated FGF Receptor 2 isoform IIIb with PTR657 | | Descriptor: | Fibroblast growth factor receptor 2, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lin, C.-C, Wieteska, L, Poncet-Montange, G, Suen, K.M, Arold, S.T, Ahmed, Z, Ladbury, J.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The combined action of the intracellular regions regulates FGFR2 kinase activity
Commun Biol, 6, 2023
|
|
8J2S
 
 | |
2QFC
 
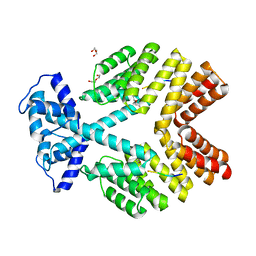 | | Crystal Structure of Bacillus thuringiensis PlcR complexed with PapR | | Descriptor: | C-terminus pentapeptide from PapR protein, DI(HYDROXYETHYL)ETHER, PlcR protein | | Authors: | Declerck, N, Chaix, D, Rugani, N, Hoh, F, Arold, S.T. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4NY0
 
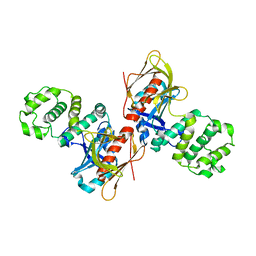 | |
1OW6
 
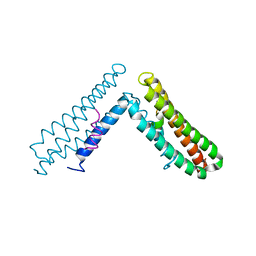 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
7C8L
 
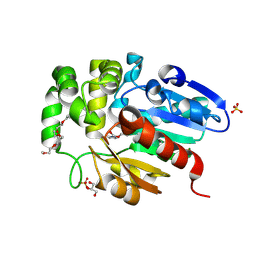 | | Hybrid designing of potent inhibitors of Striga strigolactone receptor ShHTL7 | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Shahul Hameed, U.F, Arold, S.T. | | Deposit date: | 2020-06-02 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rational design of Striga hermonthica-specific seed germination inhibitors.
Plant Physiol., 188, 2022
|
|
7D7S
 
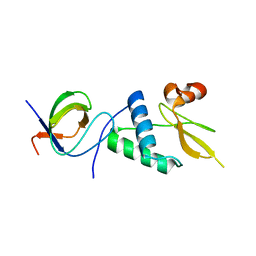 | |
6LES
 
 | |
6LF3
 
 | |
7Y7U
 
 | |
6LVW
 
 | |
7W9U
 
 | |
7W8I
 
 | |
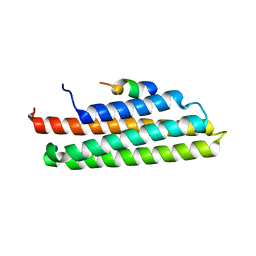
3B71
 
 | |
3H0H
 
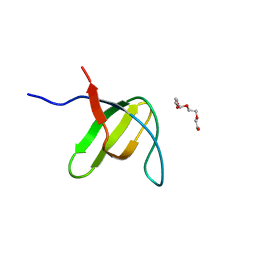 | |
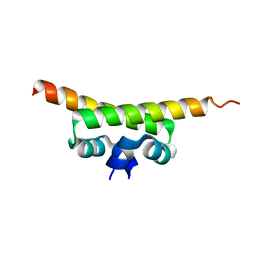
1OV9
 
 | |
3H0F
 
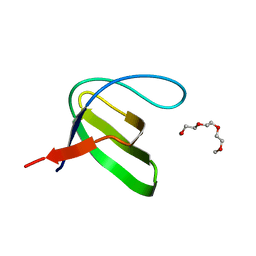 | | Crystal structure of the human Fyn SH3 R96W mutant | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Ponchon, L, Hoh, F, Labesse, G, Dumas, C, Arold, S.T. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
3H0I
 
 | |
1OW7
 
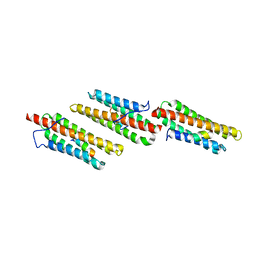 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
1OW8
 
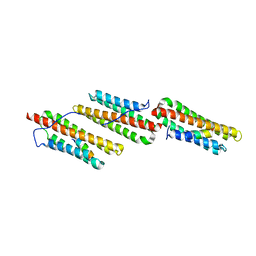 | | Paxillin LD2 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular Recognition of Paxillin LD Motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
5Z95
 
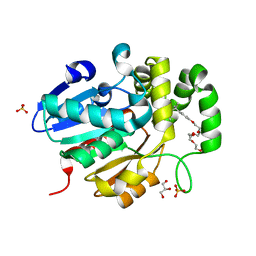 | | Structural basis for specific inhibition of highly sensitive ShHTL7 receptor | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOX Y}-ETHOXY)-ETHANOL, GLYCEROL, Hyposensitive to light 7, ... | | Authors: | Hameed, U.S, Arold, S.T. | | Deposit date: | 2018-02-02 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor.
EMBO Rep., 19, 2018
|
|
5YVS
 
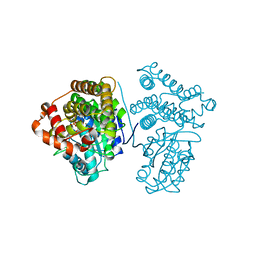 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NADP | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVR
 
 | | Crystal Structure of the H277A mutant of ADH/D1, an archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase | | Descriptor: | MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
5YVM
 
 | | Crystal Structure of the archaeal halo-thermophilic Red Sea brine pool alcohol dehydrogenase ADH/D1 bound to NZQ | | Descriptor: | 5,6-DIHYDROXY-NADP, MANGANESE (II) ION, alcohol dehydrogenase | | Authors: | Groetzinger, S.W, Strillinger, E, Frank, A, Eppinger, J, Groll, M, Arold, S.T. | | Deposit date: | 2017-11-26 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification and Experimental Characterization of an Extremophilic Brine Pool Alcohol Dehydrogenase from Single Amplified Genomes
ACS Chem. Biol., 13, 2018
|
|
