2NLU
 
 | |
5HUA
 
 | | Structure of C. glabrata FKBP12-FK506 complex | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Pathogenic Fungal FKBP12s Reveal Possible Self-Catalysis Function.
Mbio, 7, 2016
|
|
5HW7
 
 | |
5HTG
 
 | |
5HWC
 
 | |
5J6E
 
 | |
5I98
 
 | |
5GWP
 
 | | Crystal structure of RCAR3:PP2C wild-type with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.577 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
5GWO
 
 | | Crystal structure of RCAR3:PP2C S265F/I267M with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Han, S, Lee, S. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Modulation of ABA Signaling by Altering VxG Phi L Motif of PP2Cs in Oryza sativa.
Mol Plant, 10, 2017
|
|
6O5I
 
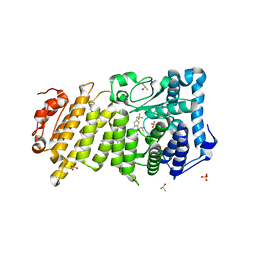 | | Menin in complex with MI-3454 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, SULFATE ION, ... | | Authors: | Linhares, B.M, Klossowski, S, Cierpicki, T, Grembecka, J. | | Deposit date: | 2019-03-03 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.24025619 Å) | | Cite: | Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia.
J.Clin.Invest., 130, 2020
|
|
5GTJ
 
 | |
4Q9J
 
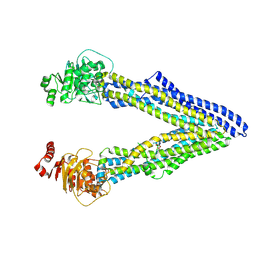 | | P-glycoprotein cocrystallised with QZ-Val | | Descriptor: | Multidrug resistance protein 1A, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9H
 
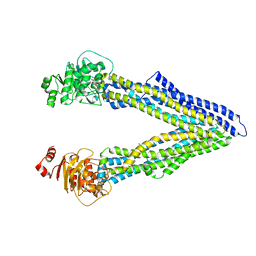 | | P-glycoprotein at 3.4 A resolution | | Descriptor: | Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9K
 
 | | P-glycoprotein cocrystallised with QZ-Leu | | Descriptor: | (30F)L(30F)L(30F)L Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5GLJ
 
 | |
4Q9L
 
 | | P-glycoprotein cocrystallised with QZ-Phe | | Descriptor: | (30F)F(30F)F(30F)F Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9I
 
 | | P-glycoprotein cocrystallised with QZ-Ala | | Descriptor: | (30F)A(30F)A(30F)A Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.781 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3BD3
 
 | |
5B8I
 
 | | Crystal structure of Calcineurin A and Calcineurin B in complex with FKBP12 and FK506 from Coccidioides immitis RS | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Dranow, D.M, Lorimer, D.D, Edwards, T.E. | | Deposit date: | 2015-05-03 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
3BD5
 
 | |
3BD4
 
 | |
4IKC
 
 | | Crystal Structure of catalytic domain of PTPRQ | | Descriptor: | CHLORIDE ION, Phosphotidylinositol phosphatase PTPRQ, SULFATE ION | | Authors: | Yu, K.R, Ryu, S.E, Kim, S.J. | | Deposit date: | 2012-12-26 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis for the dephosphorylating activity of PTPRQ towards phosphatidylinositide substrates
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3UL8
 
 | | Crystal structure of the TV3 mutant V134L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3ULA
 
 | | Crystal structure of the TV3 mutant F63W-MD-2-Eritoran complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-O-DECYL-2-DEOXY-6-O-{2-DEOXY-3-O-[(3R)-3-METHOXYDECYL]-6-O-METHYL-2-[(11Z)-OCTADEC-11-ENOYLAMINO]-4-O-PHOSPHONO-BETA-D-GLUCOPYRANOSYL}-2-[(3-OXOTETRADECANOYL)AMINO]-1-O-PHOSPHONO-ALPHA-D-GLUCOPYRANOSE, Lymphocyte antigen 96, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
3UL7
 
 | | Crystal structure of the TV3 mutant F63W | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Toll-like receptor 4, ... | | Authors: | Kim, H.J, Cheong, H.K, Jeon, Y.H. | | Deposit date: | 2011-11-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Rational Design of a Toll-like Receptor 4 (TLR4) Decoy Receptor with High Binding Affinity for a Target Protein.
Plos One, 7, 2012
|
|
