3I2U
 
 | | Crystal structure of the haiprin ribozyme with a 2',5'-linked substrate and N1-deazaadenosine at position A10 | | Descriptor: | 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
3GS1
 
 | | An all-RNA Hairpin Ribozyme with mutation A38N1dA | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3GS5
 
 | | An all-RNA hairpin ribozyme A38N1dA variant with a product mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (25-MER), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
3GS8
 
 | | An all-RNA hairpin ribozyme A38N1dA38 variant with a transition-state mimic substrate strand | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3'), ... | | Authors: | Spitale, R.C, Volpini, R, Heller, M.G, Krucinska, J, Cristalli, G, Wedekind, J.E. | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of an imino group indispensable for cleavage by a small ribozyme.
J.Am.Chem.Soc., 131, 2009
|
|
2NPZ
 
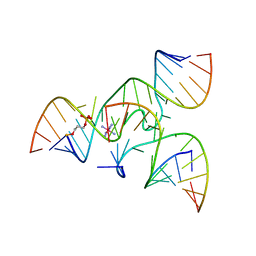 | | Crystal structure of junctioned hairpin ribozyme incorporating synthetic propyl linker | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2006-10-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2NPY
 
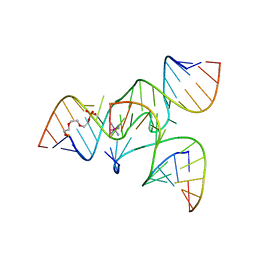 | | Crystal Structure of a junctioned hairpin ribozyme incorporating 9atom linker and 2'-deoxy 2'-amino U at A-1 | | Descriptor: | 2-[2-(2-HYDROXYETHOXY)ETHOXY]ETHYL DIHYDROGEN PHOSPHATE, 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2006-10-30 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A posteriori design of crystal contacts to improve the X-ray diffraction properties of a small RNA enzyme.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1X9K
 
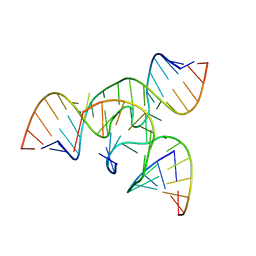 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*AP*AP*UP*AP*GP*AP*GP*AP*AP*GP*CP*GP*A)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*GP*CP*AP*GP*UP*CP*CP*UP*AP*UP*U)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-21 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
1X9C
 
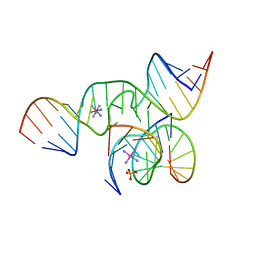 | | An all-RNA Hairpin Ribozyme with mutation U39C | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2004-08-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Conformational Heterogeneity at Position U37 of an All-RNA Hairpin Ribozyme with Implications for Metal Binding and the Catalytic Structure of the S-Turn.
Biochemistry, 44, 2005
|
|
3B5F
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating the Ade38Dap Mutation and a 2',5' Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBK
 
 | | Miminally Junctioned Hairpin Ribozyme Incorporates A38C and 2'5'-phosphodiester Linkage within Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBI
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporating A38(2AP) and A-1 2'-O-Me Modifications near Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3BBM
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38C and 2'O-Me Modification at Active Site | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-09 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B5A
 
 | | Crystal Structure of a Minimally Hinged Hairpin Ribozyme Incorporating A38G mutation with a 2'OMe modification at the active site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B91
 
 | | Minimally Hinged Hairpin Ribozyme Incorporates Ade38(2AP) and 2',5'-Phosphodiester Linkage Mutations at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-11-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B58
 
 | | Minimally Junctioned Hairpin Ribozyme Incorporates A38G Mutation and a 2',5'-Phosphodiester Linkage at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
3B5S
 
 | | Minimally Hinged Hairpin Ribozyme Incorporates A38DAP Mutation and 2'-O-methyl Modification at the Active Site | | Descriptor: | 29-mer Loop A and Loop B Ribozyme strand, COBALT HEXAMMINE(III), Loop A Substrate strand, ... | | Authors: | MacElrevey, C, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-10-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural effects of nucleobase variations at key active site residue Ade38 in the hairpin ribozyme.
Rna, 14, 2008
|
|
7MQP
 
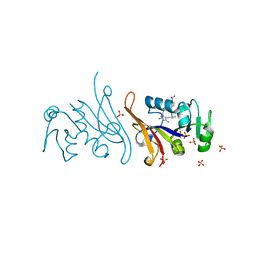 | | E. coli dihydrofolate reductase complexed with 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-4'-methyl[1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7MYL
 
 | |
7RGO
 
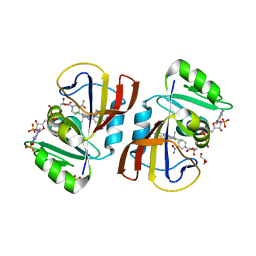 | | DfrA5 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, Dihydrofolate reductase type 5, GLYCEROL, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RGK
 
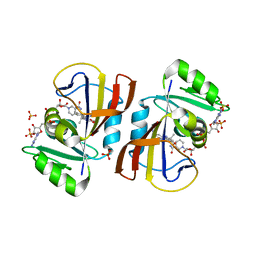 | | DfrA5 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase type 5, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RGJ
 
 | | DfrA1 complexed with NADPH and 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, 5-[(3S)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, CALCIUM ION, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7REB
 
 | | E. coli dihydrofolate reductase complexed with 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7REG
 
 | | DfrA1 complexed with NADPH and 4'-chloro-3'-(4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl)-[1,1'-biphenyl]-4-carboxamide (UCP1228) | | Descriptor: | 4'-chloro-3'-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl][1,1'-biphenyl]-4-carboxamide, CALCIUM ION, Dihydrofolate reductase type 1, ... | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6PBO
 
 | | Staphylococcus aureus Dihydrofolate reductase in complex with NADPH and UCP1232 | | Descriptor: | (4-{6-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-2H-1,3-benzodioxol-4-yl}phenyl)acetic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Toward Broad Spectrum Dihydrofolate Reductase Inhibitors Targeting Trimethoprim Resistant Enzymes Identified in Clinical Isolates of Methicillin ResistantStaphylococcus aureus.
Acs Infect Dis., 5, 2019
|
|
2FGP
 
 | | Crystal structure of a minimal, all RNA hairpin ribozyme with modifications (g8dap, u39c) at ph 8.6 | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*(N6G)P*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*AP*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Salter, J.D, Wedekind, J.E. | | Deposit date: | 2005-12-22 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Water in the Active Site of an All-RNA Hairpin Ribozyme and Effects of Gua8 Base Variants on the Geometry of Phosphoryl Transfer.
Biochemistry, 45, 2006
|
|
