7EVO
 
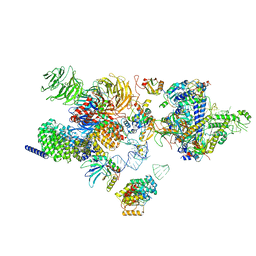 | | The cryo-EM structure of the human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
8WPU
 
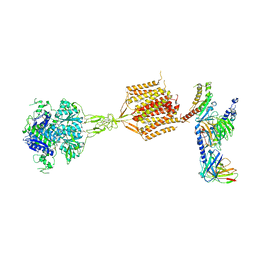 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPG
 
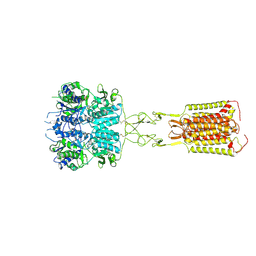 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
2JTR
 
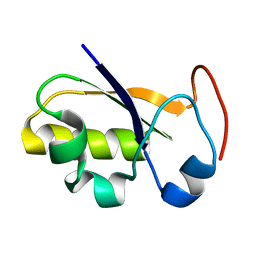 | | rhodanese persulfide from E. coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
7JVN
 
 | |
2JTQ
 
 | | Rhodanese from E.coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
2JSO
 
 | | Antimicrobial resistance protein | | Descriptor: | Polymyxin resistance protein pmrD | | Authors: | Jin, C, Fu, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | First structure of the polymyxin resistance proteins.
Biochem.Biophys.Res.Commun., 361, 2007
|
|
2JTS
 
 | | rhodanese with anions from E. coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
2K0N
 
 | |
7JVM
 
 | |
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
7FDV
 
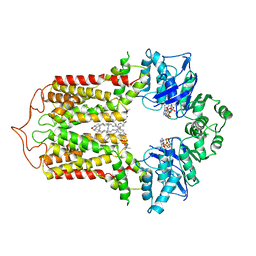 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
8H89
 
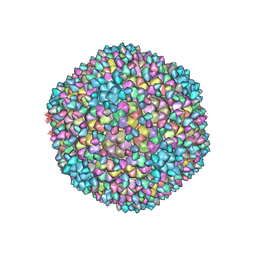 | | Capsid of Ralstonia phage GP4 | | Descriptor: | Major capsid protein, Virion associated protein | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-10-22 | | Release date: | 2022-11-16 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Capsid Structure of Ralstonia solanacearum podoviridae GP4 with a Triangulation Number T = 9.
Viruses, 14, 2022
|
|
8GVJ
 
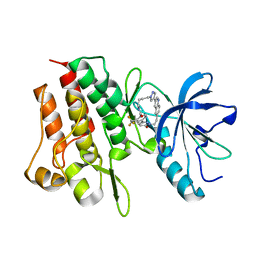 | | Crystal structure of cMET kinase domain bound by D6808 | | Descriptor: | (1^4Z,5^2E)-6^3-(trifluoromethyl)-5^1,5^6-dihydro-1^1H-8-aza-2(3,6)-quinolina-5(1,3)-pyridazina-1(4,1)-pyrazola-6(1,4)-benzenacyclododecaphane-5^6,7-dione, Hepatocyte growth factor receptor | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2022-09-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of D6808, a Highly Selective and Potent Macrocyclic c-Met Inhibitor for Gastric Cancer Harboring MET Gene Alteration Treatment.
J.Med.Chem., 65, 2022
|
|
8H3Z
 
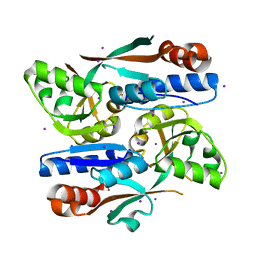 | |
8I1V
 
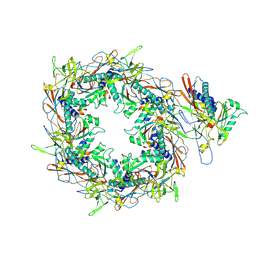 | | The asymmetric unit of P22 procapsid | | Descriptor: | Major capsid protein, Scaffolding protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8I1T
 
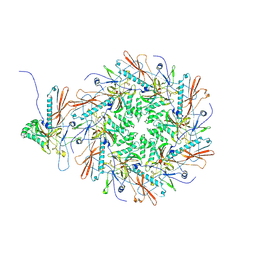 | | The asymmetric unit of P22 empty capsid | | Descriptor: | Major capsid protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
7VC9
 
 | | Tom20 subunits | | Descriptor: | Mitochondrial import receptor subunit TOM20 homolog | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VD2
 
 | | Human TOM complex without cross-linking | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VDD
 
 | | Human TOM complex with cross-linking | | Descriptor: | Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VJT
 
 | | Crystal Structure of Mtb Pks13-TE in complex with inhibitor coumestan derivative 8 | | Descriptor: | 3,8-bis(oxidanyl)-7-(piperidin-1-ylmethyl)-[1]benzofuro[3,2-c]chromen-6-one, Polyketide synthase Pks13 (Termination polyketide synthase) | | Authors: | Zhang, W, Wang, S.S, Yu, L.F. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of Coumestan Derivatives as Polyketide Synthase 13-Thioesterase(Pks13-TE) Inhibitors with Improved hERG Profiles for Mycobacterium tuberculosis Treatment.
J.Med.Chem., 65, 2022
|
|
7VPX
 
 | | The cryo-EM structure of the human pre-A complex | | Descriptor: | 5SS, DnaJ homolog subfamily C member 8, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-10-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
7CFP
 
 | |
7CFQ
 
 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | Authors: | Zhao, J, Zhang, X, Zang, J. | | Deposit date: | 2020-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
7DFB
 
 | | Crystal of Arrestin2-V2Rpp-6-7-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
