1SJ0
 
 | | Human Estrogen Receptor Alpha Ligand-binding Domain in Complex with the Antagonist Ligand 4-D | | Descriptor: | (2S,3R)-2-(4-(2-(PIPERIDIN-1-YL)ETHOXY)PHENYL)-2,3-DIHYDRO-3-(4-HYDROXYPHENYL)BENZO[B][1,4]OXATHIIN-6-OL, Estrogen receptor | | Authors: | Kim, S, Wu, J.Y, Birzin, E.T, Chan, W, Pai, L.Y, Yang, Y.T, Mosley, R.T, Fitzgerald, P.M, Sharma, N, DiNinno, F, Rohrer, S.P, Schaeffer, J.M, Hammond, M.L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Estrogen Receptor Ligands. II. Discovery of Benzoxathiins as Potent, Selective Estrogen Receptor alpha Modulators.
J.Med.Chem., 47, 2004
|
|
1SFP
 
 | | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY | | Descriptor: | ASFP | | Authors: | Romao, M.J, Kolln, I, Dias, J.M, Carvalho, A.L, Romero, A, Varela, P.F, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
2JED
 
 | | The crystal structure of the kinase domain of the protein kinase C theta in complex with NVP-XAA228 at 2.32A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(8-DIMETHYLAMINOMETHYL-6,7,8,9-TETRAHYDRO-PYRIDO[1,2-A]INDOL-10-YL)-4-(1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, PROTEIN KINASE C THETA | | Authors: | Stark, W, Bitsch, F, Berner, A, Buelens, F, Graff, P, Depersin, H, Geiser, M, Knecht, R, Rahuel, J, Rummel, G, Schlaeppi, J.M, Schmitz, R, Strauss, A, Wagner, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of the Kinase Domain of the Protein Kinase C Theta in Complex with Nvp-Xaa228
To be Published
|
|
1SIH
 
 | | AGAO in covalent complex with the inhibitor MOBA ("4-(4-methylphenoxy)-2-butyn-1-amine") | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Guss, J.M, Langley, D.B, Duff, A.P. | | Deposit date: | 2004-02-29 | | Release date: | 2004-09-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Differential Inhibition of Six Copper Amine Oxidases by a Family of 4-(Aryloxy)-2-butynamines: Evidence for a New Mode of Inactivation.
Biochemistry, 43, 2004
|
|
1UTZ
 
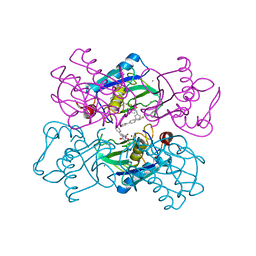 | | Crystal Structure of MMP-12 complexed to (2R)-3-({[4-[(pyridin-4-yl)phenyl]-thien-2-yl}carboxamido)(phenyl)propanoic acid | | Descriptor: | (2R)-3-({[4-[(PYRIDIN-4-YL)PHENYL]-THIEN-2-YL}CARBOXAMIDO)(PHENYL)PROPANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Novel Non-Peptidic, Non-Zinc Chelating Inhibitors Bound to Mmp-12
J.Mol.Biol., 341, 2004
|
|
1UVN
 
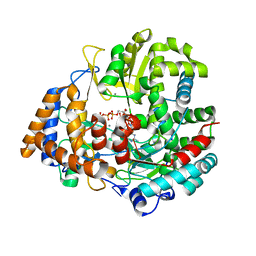 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 ca2+ inhibition complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UTT
 
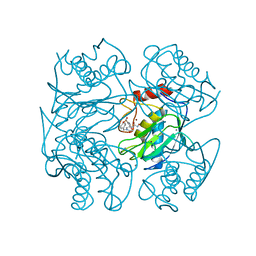 | | Crystal Structure of MMP-12 complexed to 2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl-4-(4-ethoxy[1,1-biphenyl]-4-yl)-4-oxobutanoic acid | | Descriptor: | 2-(1,3-DIOXO-1,3-DIHYDRO-2H-ISOINDOL-2-YL) ETHYL-4-(4'-ETHOXY [1,1'-BIPHENYL]-4-YL)-4-OXOBUTANOIC ACID, ACETOHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Morales, R, Perrier, S, Florent, J.M, Beltra, J, Dufour, S, De Mendez, I, Manceau, P, Tertre, A, Moreau, F, Compere, D, Dublanchet, A.C, O'Gara, M. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of novel non-peptidic, non-zinc chelating inhibitors bound to MMP-12.
J. Mol. Biol., 341, 2004
|
|
1UOS
 
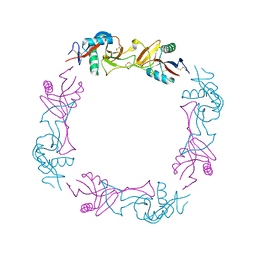 | | The Crystal Structure of the Snake Venom Toxin Convulxin | | Descriptor: | CONVULXIN ALPHA, CONVULXIN BETA | | Authors: | Batuwangala, T, Leduc, M, Gibbins, J.M, Bon, C, Jones, E.Y. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Snake-Venom Toxin Convulxin
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2LSS
 
 | | Solution structure of the R. rickettsii cold shock-like protein | | Descriptor: | Cold shock-like protein | | Authors: | Veldkamp, C.T, Peterson, F.C, Gerarden, K.P, Fuchs, A.M, Koch, J.M, Mueller, M.M. | | Deposit date: | 2012-05-04 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cold-shock-like protein from Rickettsia rickettsii.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1BB8
 
 | |
2L4Z
 
 | | NMR structure of fusion of CtIP (641-685) to LMO4-LIM1 (18-82) | | Descriptor: | DNA endonuclease RBBP8,LIM domain transcription factor LMO4, ZINC ION | | Authors: | Liew, C, Stokes, P.H, Kwan, A.H, Matthews, J.M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction of the Breast Cancer Oncogene LMO4 with the Tumour Suppressor CtIP/RBBP8.
J.Mol.Biol., 425, 2013
|
|
1BIK
 
 | | X-RAY STRUCTURE OF BIKUNIN FROM THE HUMAN INTER-ALPHA-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIKUNIN, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Guss, J.M, Ollis, D.L. | | Deposit date: | 1997-11-26 | | Release date: | 1999-03-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of bikunin from the inter-alpha-inhibitor complex: a serine protease inhibitor with two Kunitz domains.
J.Mol.Biol., 276, 1998
|
|
2L3O
 
 | | Solution structure of murine interleukin 3 | | Descriptor: | Interleukin 3 | | Authors: | Yao, S, Young, I.G, Norton, R.S, Murphy, J.M. | | Deposit date: | 2010-09-20 | | Release date: | 2011-08-10 | | Method: | SOLUTION NMR | | Cite: | Murine interleukin-3: structure, dynamics, and conformational heterogeneity in solution.
Biochemistry, 50, 2011
|
|
2L57
 
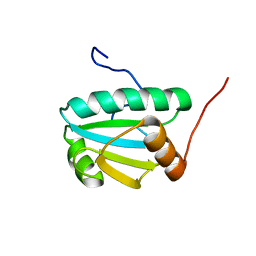 | | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Foti, R, Seidel, R.D, Bonanno, J.B, Freeman, J, Bain, K.T, Sauder, J.M, Burley, S.K, Girvin, M.E, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Uncharacterized Thioredoin-like Protein from Clostridium perfringens
To be Published
|
|
1UVL
 
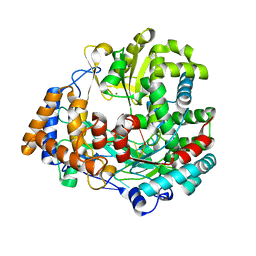 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5nt RNA. Conformation B | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
2LNH
 
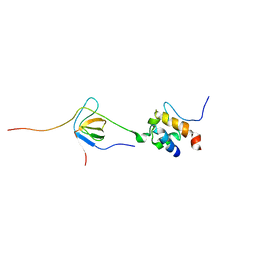 | | Enterohaemorrhagic E. coli (EHEC) exploits a tryptophan switch to hijack host F-actin assembly | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, Neural Wiskott-Aldrich syndrome protein, Secreted effector protein EspF(U) | | Authors: | Aitio, O, Hellman, M, Skehan, B, Kesti, T, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2011-12-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly.
Structure, 20, 2012
|
|
1BE7
 
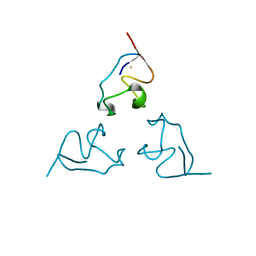 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
2LTD
 
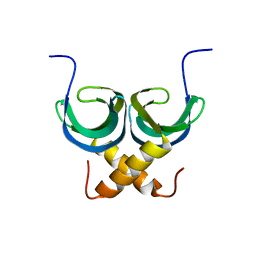 | | Solution NMR Structure of apo YdbC from Lactococcus lactis, Northeast Structural Genomics Consortium (NESG) Target KR150 | | Descriptor: | Uncharacterized protein ydbC | | Authors: | Rossi, P, Barbieri, C.M, Aramini, J.M, Bini, E, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of apo- and ssDNA-bound YdbC from Lactococcus lactis uncover the function of protein domain family DUF2128 and expand the single-stranded DNA-binding domain proteome.
Nucleic Acids Res., 41, 2013
|
|
2LA4
 
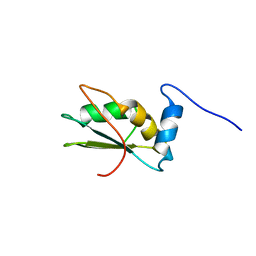 | | NMR structure of the C-terminal RRM domain of poly(U) binding 1 | | Descriptor: | Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Santiveri, C.M, Mirassou, Y, Rico-Lastres, P, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2011-03-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pub1p C-terminal RRM domain interacts with Tif4631p through a conserved region neighbouring the Pab1p binding site
Plos One, 6, 2011
|
|
1UNY
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UO3
 
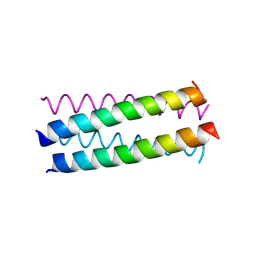 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UW7
 
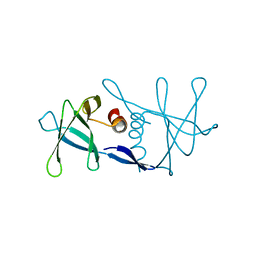 | | Nsp9 protein from SARS-coronavirus. | | Descriptor: | NSP9 | | Authors: | Sutton, G, Fry, E, Carter, L, Sainsbury, S, Walter, T, Nettleship, J, Berrow, N, Owens, R, Gilbert, R, Davidson, A, Siddell, S, Poon, L.L.M, Diprose, J, Alderton, D, Walsh, M, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Nsp9 Replicase Protein of Sars-Coronavirus, Structure and Functional Insights
Structure, 12, 2004
|
|
1BI5
 
 | | CHALCONE SYNTHASE FROM ALFALFA | | Descriptor: | CHALCONE SYNTHASE | | Authors: | Ferrer, J.L, Jez, J.M, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Nat.Struct.Biol., 6, 1999
|
|
1UO1
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UNU
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
