6K2H
 
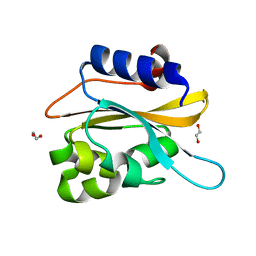 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | Descriptor: | 1,2-ETHANEDIOL, NreA | | Authors: | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
6MUR
 
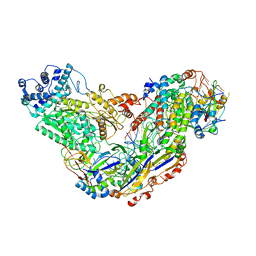 | | Cryo-EM structure of Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (27-MER), RNA (5'-R(P*GP*CP*AP*AP*AP*CP*CP*GP*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*C)-3'), Uncharacterized protein, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6MUT
 
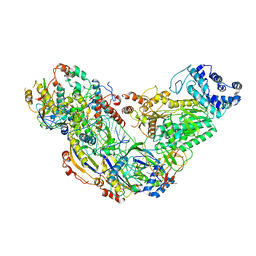 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
7XHE
 
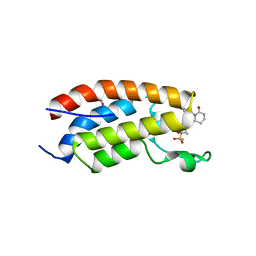 | | Crystal structure of CBP bromodomain liganded with CCS151 | | Descriptor: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
 | | Crystal structure of CBP bromodomain liganded with CCS150 | | Descriptor: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XH6
 
 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
6MRO
 
 | | Crystal structure of methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution, Northeast Structural Genomics Consortium (NESG) Target MvR53. | | Descriptor: | CALCIUM ION, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Forouhar, F, Wang, C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-10-15 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution.
To Be Published
|
|
6MUU
 
 | | Cryo-EM structure of Csm-crRNA binary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (25-MER), Uncharacterized protein Csm1, Uncharacterized protein Csm2, ... | | Authors: | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6N9N
 
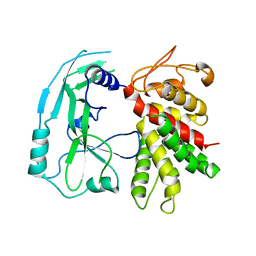 | | Crystal structure of murine GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
8Y2L
 
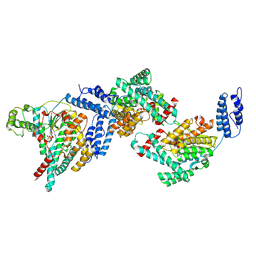 | | The Crystal Structure of Glucosyltransferase TcdB from Clostridioides difficile | | Descriptor: | Glucosyltransferase TcdB, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Fan, S, Wei, X, Lv, R, Wang, C, Tang, M, Jin, Y, Yang, Z. | | Deposit date: | 2024-01-26 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | The Crystal Structure of Glucosyltransferase TcdB from Clostridioides difficile
To Be Published
|
|
8XNA
 
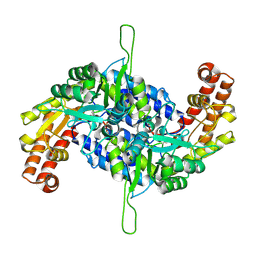 | | Crystal structure of serine hydroxymethyltransferase 2 | | Descriptor: | Serine hydroxymethyltransferase, mitochondrial | | Authors: | Fan, S, Lv, R, Wang, C, Tang, M, Wei, X, Jin, Y, Yang, Z. | | Deposit date: | 2023-12-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase 2
To Be Published
|
|
8XND
 
 | | Crystal structure of serine hydroxymethyltransferase 1 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Fan, S, Wei, X, Lv, R, Wang, C, Tang, M, Jin, Y, Yang, Z. | | Deposit date: | 2023-12-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase 1
To Be Published
|
|
2JQC
 
 | | A L-amino acid mutant of a D-amino acid containing conopeptide | | Descriptor: | L-mr12 | | Authors: | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Purification and structural characterization of a d-amino acid-containing conopeptide, conomarphin, from Conus marmoreus
Febs J., 275, 2008
|
|
7VYQ
 
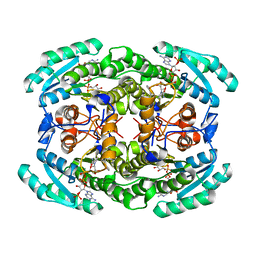 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | Descriptor: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | Authors: | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7XCE
 
 | | Crystal structure of Ankyrin G in complex with neurofascin | | Descriptor: | GLYCEROL, Neurofascin,Ankyrin-3, SULFATE ION | | Authors: | He, L, Li, J, Wang, C. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Ankyrin-G in complex with a fragment of Neurofascin reveals binding mechanisms required for integrity of the axon initial segment.
J.Biol.Chem., 298, 2022
|
|
7XNG
 
 | | Crystal structure of CBP bromodomain liganded with Y08092(31g) | | Descriptor: | 3-[(1-ethanoylindol-3-yl)carbonylamino]-5-[[(2S)-oxan-2-yl]oxymethyl]benzoic acid, CREB-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08092(31g)
To Be Published
|
|
7XM7
 
 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
2VGB
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
7DMG
 
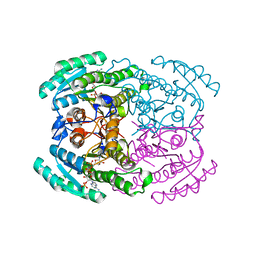 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | Descriptor: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-03 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLM
 
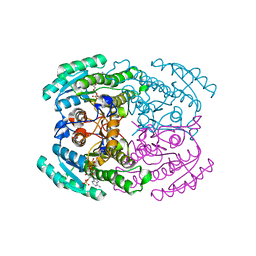 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | Descriptor: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLD
 
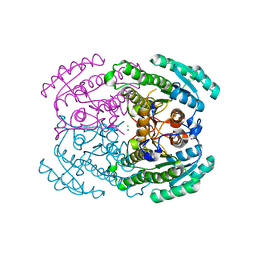 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | Descriptor: | Carbonyl Reductase, MAGNESIUM ION | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-27 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DN1
 
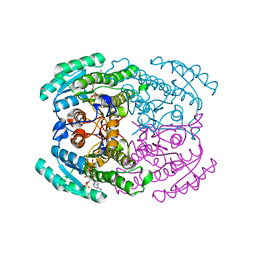 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-12-08 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | Descriptor: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2020-11-28 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7EP7
 
 | | The complex structure of Gpsm2 and Whirlin | | Descriptor: | G-protein-signaling modulator 2, Whirlin | | Authors: | Lin, L, Shi, Y, Wang, C, Zhu, J. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Promotion of row 1-specific tip complex condensates by Gpsm2-G alpha i provides insights into row identity of the tallest stereocilia.
Sci Adv, 8, 2022
|
|
